
Glycomyces sambucus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Glycomycetales; Glycomycetaceae; Glycomyces
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
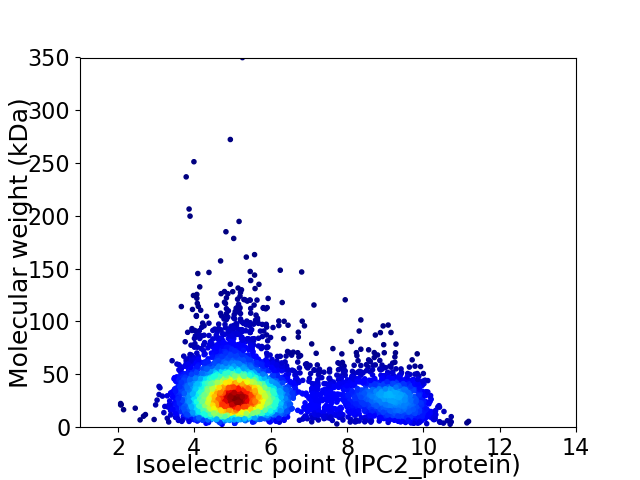
Virtual 2D-PAGE plot for 5037 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
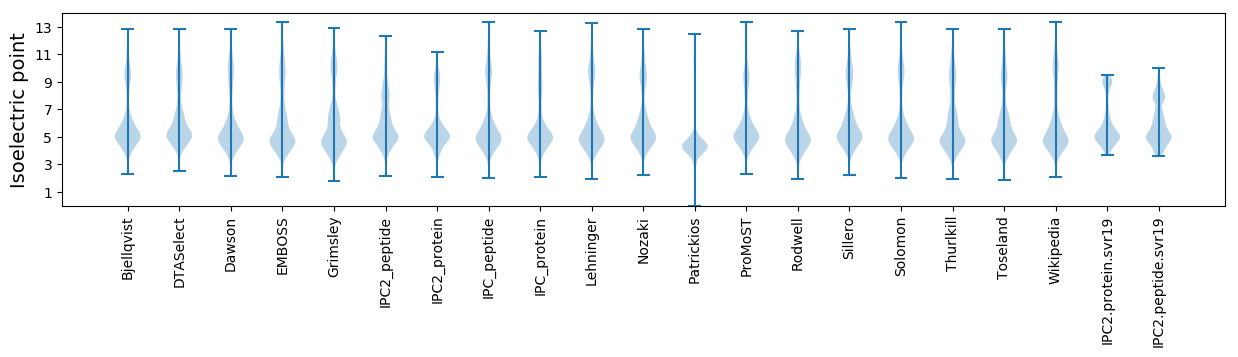
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G9CNK5|A0A1G9CNK5_9ACTN Uncharacterized protein OS=Glycomyces sambucus OX=380244 GN=SAMN05216298_0435 PE=4 SV=1
MM1 pKa = 7.09HH2 pKa = 7.45RR3 pKa = 11.84YY4 pKa = 8.44VRR6 pKa = 11.84RR7 pKa = 11.84GLVTAVMSGGLILLGSSMASADD29 pKa = 3.83EE30 pKa = 4.38GSGDD34 pKa = 3.98LLGLGGVVDD43 pKa = 4.61SVTSGLTGDD52 pKa = 4.47DD53 pKa = 3.66QGDD56 pKa = 3.93TASGDD61 pKa = 3.29ASADD65 pKa = 3.77GNSDD69 pKa = 3.09AAANGGNGGDD79 pKa = 3.83SGNAGDD85 pKa = 4.09ATSDD89 pKa = 3.35NTATSGNSGDD99 pKa = 4.31SGNSGDD105 pKa = 3.77TGGNSALNVCVLAEE119 pKa = 4.08CHH121 pKa = 6.31ASADD125 pKa = 4.07SGDD128 pKa = 3.95SGDD131 pKa = 3.92TGSTGDD137 pKa = 3.85TGDD140 pKa = 4.02AANSSHH146 pKa = 6.26TSGGNSGDD154 pKa = 3.46AGEE157 pKa = 5.22GGNADD162 pKa = 3.88AGAEE166 pKa = 4.15SGNTADD172 pKa = 5.2SGEE175 pKa = 4.3VWSEE179 pKa = 4.03TGWFTDD185 pKa = 3.36SSGDD189 pKa = 3.66TASGDD194 pKa = 3.38ASADD198 pKa = 3.57GDD200 pKa = 3.86SDD202 pKa = 3.93AAATGGNGGDD212 pKa = 3.88SGDD215 pKa = 4.35AGDD218 pKa = 4.03ATSDD222 pKa = 3.37NTATSGNSGDD232 pKa = 4.29SGDD235 pKa = 4.04SGSTGGNSALNLCVLAEE252 pKa = 4.17CHH254 pKa = 6.56ASADD258 pKa = 4.05SGNSGDD264 pKa = 3.94TGSTGDD270 pKa = 4.93TGNAWNTSHH279 pKa = 7.13TNGGHH284 pKa = 6.2SGDD287 pKa = 4.51AGDD290 pKa = 5.19GGNADD295 pKa = 4.37AEE297 pKa = 4.34AGSWNSAHH305 pKa = 7.06IGDD308 pKa = 4.22AVSDD312 pKa = 3.9NGNDD316 pKa = 2.69WWHH319 pKa = 5.53FTGQNAHH326 pKa = 5.84GWHH329 pKa = 6.91HH330 pKa = 6.34GTTGDD335 pKa = 3.87TASGDD340 pKa = 3.42ATAGGDD346 pKa = 3.4SDD348 pKa = 4.76AAATGGNGGTSGDD361 pKa = 3.92AGDD364 pKa = 3.72ATSANTATTGDD375 pKa = 4.05SGDD378 pKa = 4.1SGNSGDD384 pKa = 3.91TGGNTALGLGEE395 pKa = 4.45SDD397 pKa = 3.6QNRR400 pKa = 11.84HH401 pKa = 5.64GWHH404 pKa = 6.48HH405 pKa = 6.54WCGCWKK411 pKa = 10.42HH412 pKa = 6.91DD413 pKa = 4.12DD414 pKa = 3.84ASSTEE419 pKa = 4.16VNSGDD424 pKa = 4.08SGNTGWTGTTGDD436 pKa = 3.64AWNTSHH442 pKa = 6.87TMGGHH447 pKa = 6.23SGDD450 pKa = 3.94AGNGGDD456 pKa = 3.6AWAGAWSANTAHH468 pKa = 6.72TGWAHH473 pKa = 6.32AVV475 pKa = 3.26
MM1 pKa = 7.09HH2 pKa = 7.45RR3 pKa = 11.84YY4 pKa = 8.44VRR6 pKa = 11.84RR7 pKa = 11.84GLVTAVMSGGLILLGSSMASADD29 pKa = 3.83EE30 pKa = 4.38GSGDD34 pKa = 3.98LLGLGGVVDD43 pKa = 4.61SVTSGLTGDD52 pKa = 4.47DD53 pKa = 3.66QGDD56 pKa = 3.93TASGDD61 pKa = 3.29ASADD65 pKa = 3.77GNSDD69 pKa = 3.09AAANGGNGGDD79 pKa = 3.83SGNAGDD85 pKa = 4.09ATSDD89 pKa = 3.35NTATSGNSGDD99 pKa = 4.31SGNSGDD105 pKa = 3.77TGGNSALNVCVLAEE119 pKa = 4.08CHH121 pKa = 6.31ASADD125 pKa = 4.07SGDD128 pKa = 3.95SGDD131 pKa = 3.92TGSTGDD137 pKa = 3.85TGDD140 pKa = 4.02AANSSHH146 pKa = 6.26TSGGNSGDD154 pKa = 3.46AGEE157 pKa = 5.22GGNADD162 pKa = 3.88AGAEE166 pKa = 4.15SGNTADD172 pKa = 5.2SGEE175 pKa = 4.3VWSEE179 pKa = 4.03TGWFTDD185 pKa = 3.36SSGDD189 pKa = 3.66TASGDD194 pKa = 3.38ASADD198 pKa = 3.57GDD200 pKa = 3.86SDD202 pKa = 3.93AAATGGNGGDD212 pKa = 3.88SGDD215 pKa = 4.35AGDD218 pKa = 4.03ATSDD222 pKa = 3.37NTATSGNSGDD232 pKa = 4.29SGDD235 pKa = 4.04SGSTGGNSALNLCVLAEE252 pKa = 4.17CHH254 pKa = 6.56ASADD258 pKa = 4.05SGNSGDD264 pKa = 3.94TGSTGDD270 pKa = 4.93TGNAWNTSHH279 pKa = 7.13TNGGHH284 pKa = 6.2SGDD287 pKa = 4.51AGDD290 pKa = 5.19GGNADD295 pKa = 4.37AEE297 pKa = 4.34AGSWNSAHH305 pKa = 7.06IGDD308 pKa = 4.22AVSDD312 pKa = 3.9NGNDD316 pKa = 2.69WWHH319 pKa = 5.53FTGQNAHH326 pKa = 5.84GWHH329 pKa = 6.91HH330 pKa = 6.34GTTGDD335 pKa = 3.87TASGDD340 pKa = 3.42ATAGGDD346 pKa = 3.4SDD348 pKa = 4.76AAATGGNGGTSGDD361 pKa = 3.92AGDD364 pKa = 3.72ATSANTATTGDD375 pKa = 4.05SGDD378 pKa = 4.1SGNSGDD384 pKa = 3.91TGGNTALGLGEE395 pKa = 4.45SDD397 pKa = 3.6QNRR400 pKa = 11.84HH401 pKa = 5.64GWHH404 pKa = 6.48HH405 pKa = 6.54WCGCWKK411 pKa = 10.42HH412 pKa = 6.91DD413 pKa = 4.12DD414 pKa = 3.84ASSTEE419 pKa = 4.16VNSGDD424 pKa = 4.08SGNTGWTGTTGDD436 pKa = 3.64AWNTSHH442 pKa = 6.87TMGGHH447 pKa = 6.23SGDD450 pKa = 3.94AGNGGDD456 pKa = 3.6AWAGAWSANTAHH468 pKa = 6.72TGWAHH473 pKa = 6.32AVV475 pKa = 3.26
Molecular weight: 44.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G9GPQ1|A0A1G9GPQ1_9ACTN Glycosyltransferase involved in cell wall bisynthesis OS=Glycomyces sambucus OX=380244 GN=SAMN05216298_2340 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.95TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1656086 |
27 |
3202 |
328.8 |
35.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.397 ± 0.055 | 0.697 ± 0.009 |
6.412 ± 0.036 | 6.069 ± 0.035 |
3.084 ± 0.02 | 9.27 ± 0.032 |
2.077 ± 0.016 | 3.645 ± 0.024 |
1.982 ± 0.023 | 10.134 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.841 ± 0.015 | 1.967 ± 0.021 |
5.674 ± 0.032 | 2.592 ± 0.02 |
7.314 ± 0.045 | 4.825 ± 0.025 |
5.855 ± 0.027 | 8.304 ± 0.032 |
1.673 ± 0.015 | 2.189 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
