
Wenling crustacean virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.64
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
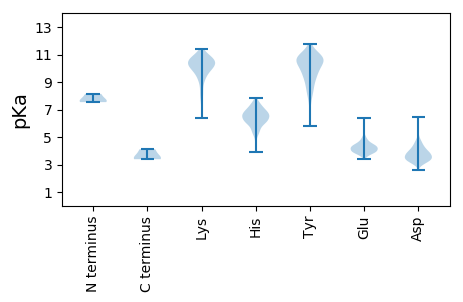
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KKE8|A0A1L3KKE8_9VIRU Uncharacterized protein OS=Wenling crustacean virus 1 OX=1923478 PE=4 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84GRR5 pKa = 11.84NIATTGRR12 pKa = 11.84GQHH15 pKa = 6.01INLKK19 pKa = 9.98GVEE22 pKa = 4.65DD23 pKa = 4.48EE24 pKa = 6.21LPDD27 pKa = 3.49IDD29 pKa = 5.58NFFDD33 pKa = 4.33YY34 pKa = 10.77ISALYY39 pKa = 10.55RR40 pKa = 11.84NAQLLFSTHH49 pKa = 4.6TTFTDD54 pKa = 3.65EE55 pKa = 4.13VAANASLNSPITRR68 pKa = 11.84FLLVPGANSLPQPGAASAFTITLLPFRR95 pKa = 11.84TSEE98 pKa = 3.86MSLVVSDD105 pKa = 4.72ISDD108 pKa = 3.69ANISDD113 pKa = 3.77QFQCPISTLQEE124 pKa = 4.41DD125 pKa = 3.84SGGWQPFTTTRR136 pKa = 11.84QPIKK140 pKa = 10.2PIAIDD145 pKa = 3.1IDD147 pKa = 3.58RR148 pKa = 11.84GRR150 pKa = 11.84EE151 pKa = 3.78HH152 pKa = 7.26LLLNVEE158 pKa = 4.31IDD160 pKa = 3.2GLTYY164 pKa = 8.4TTEE167 pKa = 3.74LTLALNSFVYY177 pKa = 9.97IHH179 pKa = 6.16VEE181 pKa = 3.88SYY183 pKa = 10.81GFSVYY188 pKa = 8.65TQSPYY193 pKa = 11.32KK194 pKa = 10.36HH195 pKa = 6.54IEE197 pKa = 3.87LAFHH201 pKa = 6.61KK202 pKa = 9.53TMQIDD207 pKa = 3.99CAAPNPTSSIHH218 pKa = 6.09NSIGTYY224 pKa = 9.52TILTILHH231 pKa = 6.05TRR233 pKa = 11.84VEE235 pKa = 4.73SPGPDD240 pKa = 3.1YY241 pKa = 10.08TQRR244 pKa = 11.84FHH246 pKa = 7.6IGSTLPFAQIDD257 pKa = 3.57AHH259 pKa = 6.79DD260 pKa = 4.39ALLTVVKK267 pKa = 10.4AVARR271 pKa = 11.84HH272 pKa = 5.62SLDD275 pKa = 3.21DD276 pKa = 4.2HH277 pKa = 6.34YY278 pKa = 11.77AEE280 pKa = 5.14AMHH283 pKa = 7.17GDD285 pKa = 3.79STLLAPLATLTQSVLALTSRR305 pKa = 11.84VPMYY309 pKa = 10.4EE310 pKa = 3.79RR311 pKa = 11.84EE312 pKa = 4.24HH313 pKa = 6.4VQFTSGVNFVTAAEE327 pKa = 4.27RR328 pKa = 11.84LWSRR332 pKa = 11.84RR333 pKa = 11.84LEE335 pKa = 4.22SNPAGGMSSSYY346 pKa = 9.54VWPGLRR352 pKa = 11.84YY353 pKa = 8.81VGKK356 pKa = 9.19PVDD359 pKa = 3.46YY360 pKa = 10.41GATYY364 pKa = 10.46FSTTHH369 pKa = 6.18TSGSCQVSIGDD380 pKa = 3.82SSLHH384 pKa = 6.52PGHH387 pKa = 6.59SSHH390 pKa = 7.83DD391 pKa = 3.74EE392 pKa = 3.78VTLQFTYY399 pKa = 10.79DD400 pKa = 3.22QDD402 pKa = 4.68FDD404 pKa = 4.77DD405 pKa = 3.69SDD407 pKa = 3.85YY408 pKa = 10.82RR409 pKa = 11.84VYY411 pKa = 10.56RR412 pKa = 11.84ARR414 pKa = 11.84YY415 pKa = 6.94TNFRR419 pKa = 11.84QFGDD423 pKa = 3.79DD424 pKa = 3.15NFFNNTFPVYY434 pKa = 9.92TSGSFSTQSVSAMNSFSAVHH454 pKa = 5.76CQGTLTFIKK463 pKa = 10.82GNTTVEE469 pKa = 4.08KK470 pKa = 10.97DD471 pKa = 3.36FDD473 pKa = 3.91TTIPIQLVRR482 pKa = 11.84DD483 pKa = 3.28GDD485 pKa = 4.06TYY487 pKa = 10.83RR488 pKa = 11.84GNWVAQIGTVSLPRR502 pKa = 11.84SNFSLPSNDD511 pKa = 2.57MWFTCWTDD519 pKa = 3.22GTVEE523 pKa = 3.96HH524 pKa = 7.57DD525 pKa = 3.35NRR527 pKa = 11.84FPFITDD533 pKa = 3.1KK534 pKa = 11.42LDD536 pKa = 3.59FGLYY540 pKa = 9.3TDD542 pKa = 4.52PPRR545 pKa = 11.84LKK547 pKa = 10.31YY548 pKa = 10.17LAAIPLIFDD557 pKa = 4.39APDD560 pKa = 3.18EE561 pKa = 4.25LLAVSDD567 pKa = 3.86ILVTGALDD575 pKa = 3.94FSLDD579 pKa = 4.02LDD581 pKa = 4.54FAPQFQPQPPFISSTEE597 pKa = 3.84NALAALTLQNSIKK610 pKa = 10.6LSIIGMDD617 pKa = 3.61LTIIDD622 pKa = 3.87NRR624 pKa = 11.84IDD626 pKa = 3.4AMEE629 pKa = 4.89SNTRR633 pKa = 11.84PNIFQFLGGIALTASMFMPMGKK655 pKa = 10.08VMLATMLAGYY665 pKa = 9.49SLEE668 pKa = 4.38TVGDD672 pKa = 3.5VMANDD677 pKa = 4.57FVTIGANTLALVATLIAHH695 pKa = 6.42HH696 pKa = 6.62RR697 pKa = 11.84MSGGVFHH704 pKa = 7.54RR705 pKa = 11.84DD706 pKa = 3.22DD707 pKa = 3.8VLSTQLPFFEE717 pKa = 6.03HH718 pKa = 7.35PDD720 pKa = 3.26LPQIEE725 pKa = 4.51NTTRR729 pKa = 11.84FRR731 pKa = 11.84TLKK734 pKa = 10.49EE735 pKa = 3.9NASLNSDD742 pKa = 3.24QFGALTRR749 pKa = 11.84PLGDD753 pKa = 3.45TTFSEE758 pKa = 4.49PFVAVRR764 pKa = 11.84TSRR767 pKa = 11.84IGVLPTPDD775 pKa = 3.99HH776 pKa = 6.64SLLPASFVDD785 pKa = 4.29AAGLLEE791 pKa = 4.4KK792 pKa = 10.57RR793 pKa = 11.84SLYY796 pKa = 9.82PEE798 pKa = 3.83HH799 pKa = 6.77HH800 pKa = 6.16QLHH803 pKa = 6.5VNTLQLYY810 pKa = 5.84EE811 pKa = 4.2TEE813 pKa = 4.6GEE815 pKa = 4.38VKK817 pKa = 10.21AVNVLTVLGVSDD829 pKa = 4.52GPQRR833 pKa = 11.84FNGVFPSASGFGHH846 pKa = 6.28TPSEE850 pKa = 4.22PGVFTLLHH858 pKa = 6.45SGTSQSSVIPLEE870 pKa = 4.34SLSGATPEE878 pKa = 4.77QILRR882 pKa = 11.84DD883 pKa = 3.87YY884 pKa = 10.51RR885 pKa = 11.84AKK887 pKa = 10.42CLSIGKK893 pKa = 9.9SYY895 pKa = 11.01IDD897 pKa = 4.09TEE899 pKa = 4.19NADD902 pKa = 3.95LDD904 pKa = 4.16TLRR907 pKa = 11.84RR908 pKa = 11.84WYY910 pKa = 10.57LGSNPARR917 pKa = 11.84PLHH920 pKa = 5.21THH922 pKa = 6.94LPTFTGTAKK931 pKa = 10.29SAQVNIPIEE940 pKa = 4.2LVEE943 pKa = 4.53QITASMADD951 pKa = 3.32HH952 pKa = 6.51NAFGKK957 pKa = 9.32PYY959 pKa = 10.43KK960 pKa = 10.82YY961 pKa = 10.9NLTNNNCKK969 pKa = 10.21HH970 pKa = 6.0FADD973 pKa = 4.83SMFSMIAYY981 pKa = 9.7GEE983 pKa = 3.97RR984 pKa = 11.84DD985 pKa = 3.47EE986 pKa = 4.72MNAAFLSRR994 pKa = 11.84LSSTRR999 pKa = 11.84FNPGVITEE1007 pKa = 4.58FEE1009 pKa = 4.77LIQYY1013 pKa = 10.66LDD1015 pKa = 5.17DD1016 pKa = 4.56ININTINTVKK1026 pKa = 10.42PGGPIIYY1033 pKa = 8.95TRR1035 pKa = 11.84EE1036 pKa = 3.58GWLL1039 pKa = 3.39
MM1 pKa = 7.54PRR3 pKa = 11.84GRR5 pKa = 11.84NIATTGRR12 pKa = 11.84GQHH15 pKa = 6.01INLKK19 pKa = 9.98GVEE22 pKa = 4.65DD23 pKa = 4.48EE24 pKa = 6.21LPDD27 pKa = 3.49IDD29 pKa = 5.58NFFDD33 pKa = 4.33YY34 pKa = 10.77ISALYY39 pKa = 10.55RR40 pKa = 11.84NAQLLFSTHH49 pKa = 4.6TTFTDD54 pKa = 3.65EE55 pKa = 4.13VAANASLNSPITRR68 pKa = 11.84FLLVPGANSLPQPGAASAFTITLLPFRR95 pKa = 11.84TSEE98 pKa = 3.86MSLVVSDD105 pKa = 4.72ISDD108 pKa = 3.69ANISDD113 pKa = 3.77QFQCPISTLQEE124 pKa = 4.41DD125 pKa = 3.84SGGWQPFTTTRR136 pKa = 11.84QPIKK140 pKa = 10.2PIAIDD145 pKa = 3.1IDD147 pKa = 3.58RR148 pKa = 11.84GRR150 pKa = 11.84EE151 pKa = 3.78HH152 pKa = 7.26LLLNVEE158 pKa = 4.31IDD160 pKa = 3.2GLTYY164 pKa = 8.4TTEE167 pKa = 3.74LTLALNSFVYY177 pKa = 9.97IHH179 pKa = 6.16VEE181 pKa = 3.88SYY183 pKa = 10.81GFSVYY188 pKa = 8.65TQSPYY193 pKa = 11.32KK194 pKa = 10.36HH195 pKa = 6.54IEE197 pKa = 3.87LAFHH201 pKa = 6.61KK202 pKa = 9.53TMQIDD207 pKa = 3.99CAAPNPTSSIHH218 pKa = 6.09NSIGTYY224 pKa = 9.52TILTILHH231 pKa = 6.05TRR233 pKa = 11.84VEE235 pKa = 4.73SPGPDD240 pKa = 3.1YY241 pKa = 10.08TQRR244 pKa = 11.84FHH246 pKa = 7.6IGSTLPFAQIDD257 pKa = 3.57AHH259 pKa = 6.79DD260 pKa = 4.39ALLTVVKK267 pKa = 10.4AVARR271 pKa = 11.84HH272 pKa = 5.62SLDD275 pKa = 3.21DD276 pKa = 4.2HH277 pKa = 6.34YY278 pKa = 11.77AEE280 pKa = 5.14AMHH283 pKa = 7.17GDD285 pKa = 3.79STLLAPLATLTQSVLALTSRR305 pKa = 11.84VPMYY309 pKa = 10.4EE310 pKa = 3.79RR311 pKa = 11.84EE312 pKa = 4.24HH313 pKa = 6.4VQFTSGVNFVTAAEE327 pKa = 4.27RR328 pKa = 11.84LWSRR332 pKa = 11.84RR333 pKa = 11.84LEE335 pKa = 4.22SNPAGGMSSSYY346 pKa = 9.54VWPGLRR352 pKa = 11.84YY353 pKa = 8.81VGKK356 pKa = 9.19PVDD359 pKa = 3.46YY360 pKa = 10.41GATYY364 pKa = 10.46FSTTHH369 pKa = 6.18TSGSCQVSIGDD380 pKa = 3.82SSLHH384 pKa = 6.52PGHH387 pKa = 6.59SSHH390 pKa = 7.83DD391 pKa = 3.74EE392 pKa = 3.78VTLQFTYY399 pKa = 10.79DD400 pKa = 3.22QDD402 pKa = 4.68FDD404 pKa = 4.77DD405 pKa = 3.69SDD407 pKa = 3.85YY408 pKa = 10.82RR409 pKa = 11.84VYY411 pKa = 10.56RR412 pKa = 11.84ARR414 pKa = 11.84YY415 pKa = 6.94TNFRR419 pKa = 11.84QFGDD423 pKa = 3.79DD424 pKa = 3.15NFFNNTFPVYY434 pKa = 9.92TSGSFSTQSVSAMNSFSAVHH454 pKa = 5.76CQGTLTFIKK463 pKa = 10.82GNTTVEE469 pKa = 4.08KK470 pKa = 10.97DD471 pKa = 3.36FDD473 pKa = 3.91TTIPIQLVRR482 pKa = 11.84DD483 pKa = 3.28GDD485 pKa = 4.06TYY487 pKa = 10.83RR488 pKa = 11.84GNWVAQIGTVSLPRR502 pKa = 11.84SNFSLPSNDD511 pKa = 2.57MWFTCWTDD519 pKa = 3.22GTVEE523 pKa = 3.96HH524 pKa = 7.57DD525 pKa = 3.35NRR527 pKa = 11.84FPFITDD533 pKa = 3.1KK534 pKa = 11.42LDD536 pKa = 3.59FGLYY540 pKa = 9.3TDD542 pKa = 4.52PPRR545 pKa = 11.84LKK547 pKa = 10.31YY548 pKa = 10.17LAAIPLIFDD557 pKa = 4.39APDD560 pKa = 3.18EE561 pKa = 4.25LLAVSDD567 pKa = 3.86ILVTGALDD575 pKa = 3.94FSLDD579 pKa = 4.02LDD581 pKa = 4.54FAPQFQPQPPFISSTEE597 pKa = 3.84NALAALTLQNSIKK610 pKa = 10.6LSIIGMDD617 pKa = 3.61LTIIDD622 pKa = 3.87NRR624 pKa = 11.84IDD626 pKa = 3.4AMEE629 pKa = 4.89SNTRR633 pKa = 11.84PNIFQFLGGIALTASMFMPMGKK655 pKa = 10.08VMLATMLAGYY665 pKa = 9.49SLEE668 pKa = 4.38TVGDD672 pKa = 3.5VMANDD677 pKa = 4.57FVTIGANTLALVATLIAHH695 pKa = 6.42HH696 pKa = 6.62RR697 pKa = 11.84MSGGVFHH704 pKa = 7.54RR705 pKa = 11.84DD706 pKa = 3.22DD707 pKa = 3.8VLSTQLPFFEE717 pKa = 6.03HH718 pKa = 7.35PDD720 pKa = 3.26LPQIEE725 pKa = 4.51NTTRR729 pKa = 11.84FRR731 pKa = 11.84TLKK734 pKa = 10.49EE735 pKa = 3.9NASLNSDD742 pKa = 3.24QFGALTRR749 pKa = 11.84PLGDD753 pKa = 3.45TTFSEE758 pKa = 4.49PFVAVRR764 pKa = 11.84TSRR767 pKa = 11.84IGVLPTPDD775 pKa = 3.99HH776 pKa = 6.64SLLPASFVDD785 pKa = 4.29AAGLLEE791 pKa = 4.4KK792 pKa = 10.57RR793 pKa = 11.84SLYY796 pKa = 9.82PEE798 pKa = 3.83HH799 pKa = 6.77HH800 pKa = 6.16QLHH803 pKa = 6.5VNTLQLYY810 pKa = 5.84EE811 pKa = 4.2TEE813 pKa = 4.6GEE815 pKa = 4.38VKK817 pKa = 10.21AVNVLTVLGVSDD829 pKa = 4.52GPQRR833 pKa = 11.84FNGVFPSASGFGHH846 pKa = 6.28TPSEE850 pKa = 4.22PGVFTLLHH858 pKa = 6.45SGTSQSSVIPLEE870 pKa = 4.34SLSGATPEE878 pKa = 4.77QILRR882 pKa = 11.84DD883 pKa = 3.87YY884 pKa = 10.51RR885 pKa = 11.84AKK887 pKa = 10.42CLSIGKK893 pKa = 9.9SYY895 pKa = 11.01IDD897 pKa = 4.09TEE899 pKa = 4.19NADD902 pKa = 3.95LDD904 pKa = 4.16TLRR907 pKa = 11.84RR908 pKa = 11.84WYY910 pKa = 10.57LGSNPARR917 pKa = 11.84PLHH920 pKa = 5.21THH922 pKa = 6.94LPTFTGTAKK931 pKa = 10.29SAQVNIPIEE940 pKa = 4.2LVEE943 pKa = 4.53QITASMADD951 pKa = 3.32HH952 pKa = 6.51NAFGKK957 pKa = 9.32PYY959 pKa = 10.43KK960 pKa = 10.82YY961 pKa = 10.9NLTNNNCKK969 pKa = 10.21HH970 pKa = 6.0FADD973 pKa = 4.83SMFSMIAYY981 pKa = 9.7GEE983 pKa = 3.97RR984 pKa = 11.84DD985 pKa = 3.47EE986 pKa = 4.72MNAAFLSRR994 pKa = 11.84LSSTRR999 pKa = 11.84FNPGVITEE1007 pKa = 4.58FEE1009 pKa = 4.77LIQYY1013 pKa = 10.66LDD1015 pKa = 5.17DD1016 pKa = 4.56ININTINTVKK1026 pKa = 10.42PGGPIIYY1033 pKa = 8.95TRR1035 pKa = 11.84EE1036 pKa = 3.58GWLL1039 pKa = 3.39
Molecular weight: 114.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KKE8|A0A1L3KKE8_9VIRU Uncharacterized protein OS=Wenling crustacean virus 1 OX=1923478 PE=4 SV=1
MM1 pKa = 7.64SEE3 pKa = 4.3PFQVSEE9 pKa = 4.22VTSAKK14 pKa = 8.95GTRR17 pKa = 11.84NAPGYY22 pKa = 8.47STLDD26 pKa = 3.51PQPVQVAKK34 pKa = 10.35PVTTTYY40 pKa = 10.81NYY42 pKa = 10.59RR43 pKa = 11.84FAAPNIVFRR52 pKa = 11.84GAPGTSVLNLRR63 pKa = 11.84LSLNHH68 pKa = 6.64LKK70 pKa = 10.93LSDD73 pKa = 3.7PVLAKK78 pKa = 10.63LATSYY83 pKa = 7.87QHH85 pKa = 6.09WRR87 pKa = 11.84INNMSVEE94 pKa = 4.42TKK96 pKa = 10.66AVGAMATSSGSFYY109 pKa = 10.53TSYY112 pKa = 10.9ISDD115 pKa = 4.1PLMVVPPDD123 pKa = 3.99PEE125 pKa = 5.61DD126 pKa = 3.56LLKK129 pKa = 11.23LLVRR133 pKa = 11.84RR134 pKa = 11.84PEE136 pKa = 4.38TEE138 pKa = 3.56QHH140 pKa = 5.25TARR143 pKa = 11.84QNDD146 pKa = 3.74LQKK149 pKa = 10.33IHH151 pKa = 7.28PLGTAPSFQKK161 pKa = 10.58KK162 pKa = 6.77YY163 pKa = 9.21TYY165 pKa = 10.12TSLGGDD171 pKa = 3.64PRR173 pKa = 11.84LSEE176 pKa = 3.94YY177 pKa = 9.33GTYY180 pKa = 10.07VMAMATAPGAADD192 pKa = 3.33QMLVTVTISLEE203 pKa = 4.0IEE205 pKa = 4.23FEE207 pKa = 4.44LPTIIPDD214 pKa = 3.57TMTLSTPDD222 pKa = 3.65IWTMAQDD229 pKa = 3.48PQYY232 pKa = 11.63NFGPLGAYY240 pKa = 9.45IEE242 pKa = 4.48IRR244 pKa = 11.84THH246 pKa = 6.11SLAVPQVAGRR256 pKa = 11.84FIATPPNTIQLLLSDD271 pKa = 4.5GDD273 pKa = 3.76VSEE276 pKa = 4.74LVVVTITEE284 pKa = 3.88CDD286 pKa = 3.39AVVDD290 pKa = 3.69SSGRR294 pKa = 11.84VTLSIPLPATSNAYY308 pKa = 10.42VDD310 pKa = 3.99MPDD313 pKa = 3.0TVTPVGDD320 pKa = 3.74LEE322 pKa = 4.26IYY324 pKa = 10.66GYY326 pKa = 10.53INYY329 pKa = 9.82SEE331 pKa = 4.69DD332 pKa = 3.29RR333 pKa = 11.84SAEE336 pKa = 4.12RR337 pKa = 11.84IRR339 pKa = 11.84GMISKK344 pKa = 10.1RR345 pKa = 11.84NQSTKK350 pKa = 10.33RR351 pKa = 11.84FASTLRR357 pKa = 11.84NVNLHH362 pKa = 6.77PKK364 pKa = 8.64TKK366 pKa = 8.78TAIISTHH373 pKa = 7.34AINKK377 pKa = 9.13HH378 pKa = 5.62PEE380 pKa = 3.97NGPAQPVRR388 pKa = 11.84PP389 pKa = 4.11
MM1 pKa = 7.64SEE3 pKa = 4.3PFQVSEE9 pKa = 4.22VTSAKK14 pKa = 8.95GTRR17 pKa = 11.84NAPGYY22 pKa = 8.47STLDD26 pKa = 3.51PQPVQVAKK34 pKa = 10.35PVTTTYY40 pKa = 10.81NYY42 pKa = 10.59RR43 pKa = 11.84FAAPNIVFRR52 pKa = 11.84GAPGTSVLNLRR63 pKa = 11.84LSLNHH68 pKa = 6.64LKK70 pKa = 10.93LSDD73 pKa = 3.7PVLAKK78 pKa = 10.63LATSYY83 pKa = 7.87QHH85 pKa = 6.09WRR87 pKa = 11.84INNMSVEE94 pKa = 4.42TKK96 pKa = 10.66AVGAMATSSGSFYY109 pKa = 10.53TSYY112 pKa = 10.9ISDD115 pKa = 4.1PLMVVPPDD123 pKa = 3.99PEE125 pKa = 5.61DD126 pKa = 3.56LLKK129 pKa = 11.23LLVRR133 pKa = 11.84RR134 pKa = 11.84PEE136 pKa = 4.38TEE138 pKa = 3.56QHH140 pKa = 5.25TARR143 pKa = 11.84QNDD146 pKa = 3.74LQKK149 pKa = 10.33IHH151 pKa = 7.28PLGTAPSFQKK161 pKa = 10.58KK162 pKa = 6.77YY163 pKa = 9.21TYY165 pKa = 10.12TSLGGDD171 pKa = 3.64PRR173 pKa = 11.84LSEE176 pKa = 3.94YY177 pKa = 9.33GTYY180 pKa = 10.07VMAMATAPGAADD192 pKa = 3.33QMLVTVTISLEE203 pKa = 4.0IEE205 pKa = 4.23FEE207 pKa = 4.44LPTIIPDD214 pKa = 3.57TMTLSTPDD222 pKa = 3.65IWTMAQDD229 pKa = 3.48PQYY232 pKa = 11.63NFGPLGAYY240 pKa = 9.45IEE242 pKa = 4.48IRR244 pKa = 11.84THH246 pKa = 6.11SLAVPQVAGRR256 pKa = 11.84FIATPPNTIQLLLSDD271 pKa = 4.5GDD273 pKa = 3.76VSEE276 pKa = 4.74LVVVTITEE284 pKa = 3.88CDD286 pKa = 3.39AVVDD290 pKa = 3.69SSGRR294 pKa = 11.84VTLSIPLPATSNAYY308 pKa = 10.42VDD310 pKa = 3.99MPDD313 pKa = 3.0TVTPVGDD320 pKa = 3.74LEE322 pKa = 4.26IYY324 pKa = 10.66GYY326 pKa = 10.53INYY329 pKa = 9.82SEE331 pKa = 4.69DD332 pKa = 3.29RR333 pKa = 11.84SAEE336 pKa = 4.12RR337 pKa = 11.84IRR339 pKa = 11.84GMISKK344 pKa = 10.1RR345 pKa = 11.84NQSTKK350 pKa = 10.33RR351 pKa = 11.84FASTLRR357 pKa = 11.84NVNLHH362 pKa = 6.77PKK364 pKa = 8.64TKK366 pKa = 8.78TAIISTHH373 pKa = 7.34AINKK377 pKa = 9.13HH378 pKa = 5.62PEE380 pKa = 3.97NGPAQPVRR388 pKa = 11.84PP389 pKa = 4.11
Molecular weight: 42.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3268 |
389 |
1840 |
1089.3 |
121.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.436 ± 0.172 | 1.561 ± 0.628 |
5.569 ± 0.396 | 5.906 ± 1.041 |
4.284 ± 0.771 | 6.028 ± 0.233 |
4.131 ± 0.793 | 5.936 ± 0.124 |
3.733 ± 0.771 | 9.027 ± 0.354 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.142 ± 0.178 | 4.345 ± 0.222 |
5.508 ± 0.953 | 3.856 ± 0.093 |
5.416 ± 0.485 | 6.916 ± 0.993 |
8.048 ± 0.93 | 5.845 ± 0.487 |
0.826 ± 0.097 | 3.488 ± 0.169 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
