
Alkalihalobacillus lonarensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
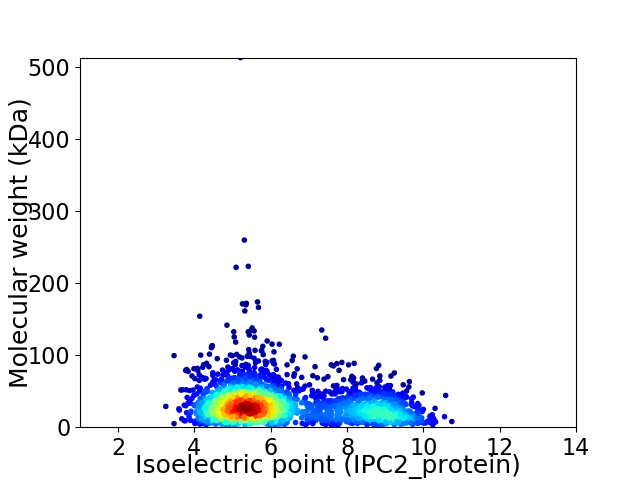
Virtual 2D-PAGE plot for 2581 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6JIM0|A0A1G6JIM0_9BACI RNA polymerase sigma factor OS=Alkalihalobacillus lonarensis OX=1464122 GN=SAMN05421737_1065 PE=3 SV=1
MM1 pKa = 7.26SQPNMPNFTPNITLDD16 pKa = 3.51RR17 pKa = 11.84DD18 pKa = 3.92DD19 pKa = 4.35VVNLLLASIAMEE31 pKa = 4.19EE32 pKa = 4.16MGLAHH37 pKa = 7.7IINAEE42 pKa = 4.05GEE44 pKa = 4.34KK45 pKa = 9.94IQYY48 pKa = 11.05ALGTLNGLTGPPATLQEE65 pKa = 3.76ILAINEE71 pKa = 4.11SAADD75 pKa = 3.65MLDD78 pKa = 3.52TIFKK82 pKa = 10.57KK83 pKa = 10.62EE84 pKa = 4.04IILEE88 pKa = 4.29SKK90 pKa = 9.82MKK92 pKa = 10.38AATNIPTLIGPTGATGATGPAGSVTNINGIFPDD125 pKa = 4.02GAGTVTITIGDD136 pKa = 3.63IPGGIEE142 pKa = 4.17NINGITAAPDD152 pKa = 3.48GSLTLDD158 pKa = 3.97AEE160 pKa = 4.66DD161 pKa = 3.89VGAFPYY167 pKa = 10.85EE168 pKa = 4.19EE169 pKa = 4.44VPSGSDD175 pKa = 2.97LDD177 pKa = 4.22SFITEE182 pKa = 4.15GVYY185 pKa = 10.55SSEE188 pKa = 4.14GNAGGPVNGPLGFTSPIWTLYY209 pKa = 10.54VSVITPGPTTSILQLLIGNPTIYY232 pKa = 10.33VRR234 pKa = 11.84SATDD238 pKa = 3.26DD239 pKa = 3.43TGTTWNPWEE248 pKa = 4.1EE249 pKa = 4.84LGTQGPTGPTGAGVTGATGATGATGATGATGATGVTGVTGVTGATGVTGATGATGVTGVTGATGVTGATGATGATGVTGVTGATGVTGATGATGVTGATGVTGATGVTGPTGVTGATGPTGPTGPTGSTGPTGATGPTGPTGPTGSTGPTGPTGVTGPTGVTGPTGPTGVTGSTGVTGITGVTGPTGPTGVTGPTGPTGVTGATGVTGVTGATGPTGSTGPTGATGPTGPDD480 pKa = 3.14PTVNEE485 pKa = 4.07VFISNSGQHH494 pKa = 5.37SVYY497 pKa = 10.38IIDD500 pKa = 5.59GNTQDD505 pKa = 4.1ILATIDD511 pKa = 3.28TTGTDD516 pKa = 3.15TRR518 pKa = 11.84GLVVNPNTNFLYY530 pKa = 10.55AINQGSSNVSVIDD543 pKa = 3.83LTTQAISTIISVAANPYY560 pKa = 10.08GLGINVDD567 pKa = 3.72TNLLYY572 pKa = 9.63VTHH575 pKa = 7.4RR576 pKa = 11.84APTNQVSVIEE586 pKa = 4.42GLNHH590 pKa = 5.22TVVTAFNIGSDD601 pKa = 3.55PVANPTVDD609 pKa = 3.3PNTNLVYY616 pKa = 10.57CANNVSNNVTVIDD629 pKa = 4.72GLTQTITATVTVGTNPFRR647 pKa = 11.84IVANPSTNQVYY658 pKa = 10.04VSNQGSDD665 pKa = 3.03NVSVINATNQTLMPTIPLAGGAAPQGITVNPNTNLIYY702 pKa = 10.81VVATNLQSVLVVDD715 pKa = 5.61WITLDD720 pKa = 3.89TIATIAVGTGAYY732 pKa = 7.47TAGANPRR739 pKa = 11.84TNLIYY744 pKa = 9.24VTKK747 pKa = 10.69VADD750 pKa = 3.77NNISVIDD757 pKa = 4.56GINNNIIATITSALPATGFRR777 pKa = 11.84SVSVLEE783 pKa = 4.06GLRR786 pKa = 11.84LTGPLYY792 pKa = 10.94SS793 pKa = 3.95
MM1 pKa = 7.26SQPNMPNFTPNITLDD16 pKa = 3.51RR17 pKa = 11.84DD18 pKa = 3.92DD19 pKa = 4.35VVNLLLASIAMEE31 pKa = 4.19EE32 pKa = 4.16MGLAHH37 pKa = 7.7IINAEE42 pKa = 4.05GEE44 pKa = 4.34KK45 pKa = 9.94IQYY48 pKa = 11.05ALGTLNGLTGPPATLQEE65 pKa = 3.76ILAINEE71 pKa = 4.11SAADD75 pKa = 3.65MLDD78 pKa = 3.52TIFKK82 pKa = 10.57KK83 pKa = 10.62EE84 pKa = 4.04IILEE88 pKa = 4.29SKK90 pKa = 9.82MKK92 pKa = 10.38AATNIPTLIGPTGATGATGPAGSVTNINGIFPDD125 pKa = 4.02GAGTVTITIGDD136 pKa = 3.63IPGGIEE142 pKa = 4.17NINGITAAPDD152 pKa = 3.48GSLTLDD158 pKa = 3.97AEE160 pKa = 4.66DD161 pKa = 3.89VGAFPYY167 pKa = 10.85EE168 pKa = 4.19EE169 pKa = 4.44VPSGSDD175 pKa = 2.97LDD177 pKa = 4.22SFITEE182 pKa = 4.15GVYY185 pKa = 10.55SSEE188 pKa = 4.14GNAGGPVNGPLGFTSPIWTLYY209 pKa = 10.54VSVITPGPTTSILQLLIGNPTIYY232 pKa = 10.33VRR234 pKa = 11.84SATDD238 pKa = 3.26DD239 pKa = 3.43TGTTWNPWEE248 pKa = 4.1EE249 pKa = 4.84LGTQGPTGPTGAGVTGATGATGATGATGATGATGVTGVTGVTGATGVTGATGATGVTGVTGATGVTGATGATGATGVTGVTGATGVTGATGATGVTGATGVTGATGVTGPTGVTGATGPTGPTGPTGSTGPTGATGPTGPTGPTGSTGPTGPTGVTGPTGVTGPTGPTGVTGSTGVTGITGVTGPTGPTGVTGPTGPTGVTGATGVTGVTGATGPTGSTGPTGATGPTGPDD480 pKa = 3.14PTVNEE485 pKa = 4.07VFISNSGQHH494 pKa = 5.37SVYY497 pKa = 10.38IIDD500 pKa = 5.59GNTQDD505 pKa = 4.1ILATIDD511 pKa = 3.28TTGTDD516 pKa = 3.15TRR518 pKa = 11.84GLVVNPNTNFLYY530 pKa = 10.55AINQGSSNVSVIDD543 pKa = 3.83LTTQAISTIISVAANPYY560 pKa = 10.08GLGINVDD567 pKa = 3.72TNLLYY572 pKa = 9.63VTHH575 pKa = 7.4RR576 pKa = 11.84APTNQVSVIEE586 pKa = 4.42GLNHH590 pKa = 5.22TVVTAFNIGSDD601 pKa = 3.55PVANPTVDD609 pKa = 3.3PNTNLVYY616 pKa = 10.57CANNVSNNVTVIDD629 pKa = 4.72GLTQTITATVTVGTNPFRR647 pKa = 11.84IVANPSTNQVYY658 pKa = 10.04VSNQGSDD665 pKa = 3.03NVSVINATNQTLMPTIPLAGGAAPQGITVNPNTNLIYY702 pKa = 10.81VVATNLQSVLVVDD715 pKa = 5.61WITLDD720 pKa = 3.89TIATIAVGTGAYY732 pKa = 7.47TAGANPRR739 pKa = 11.84TNLIYY744 pKa = 9.24VTKK747 pKa = 10.69VADD750 pKa = 3.77NNISVIDD757 pKa = 4.56GINNNIIATITSALPATGFRR777 pKa = 11.84SVSVLEE783 pKa = 4.06GLRR786 pKa = 11.84LTGPLYY792 pKa = 10.94SS793 pKa = 3.95
Molecular weight: 77.55 kDa
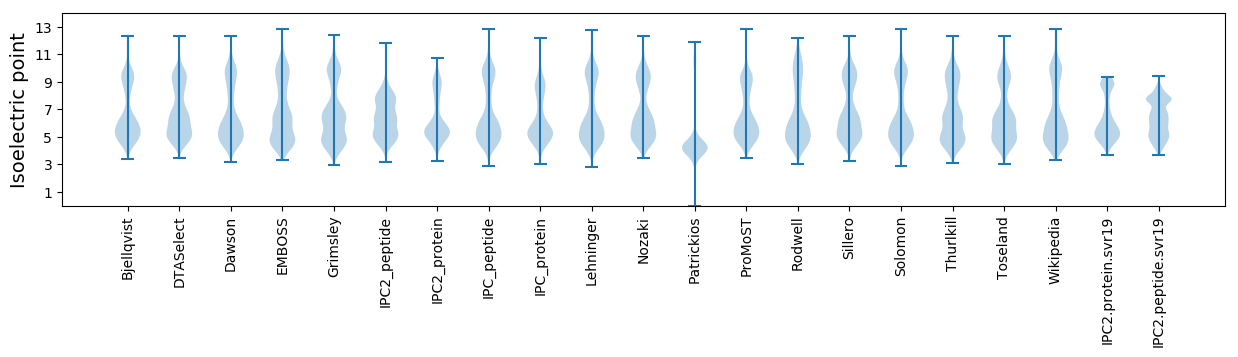
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6NC61|A0A1G6NC61_9BACI Acetyltransferase (GNAT) domain-containing protein OS=Alkalihalobacillus lonarensis OX=1464122 GN=SAMN05421737_11232 PE=4 SV=1
MM1 pKa = 7.65PNISRR6 pKa = 11.84IQALCRR12 pKa = 11.84MMMGLTLLAWAAARR26 pKa = 11.84FTRR29 pKa = 11.84RR30 pKa = 11.84PYY32 pKa = 10.85CPFSFIMALFGAQKK46 pKa = 8.89TAEE49 pKa = 4.13GTLRR53 pKa = 11.84FCPLVALRR61 pKa = 11.84KK62 pKa = 9.25RR63 pKa = 11.84CYY65 pKa = 10.33EE66 pKa = 3.84RR67 pKa = 11.84DD68 pKa = 3.31
MM1 pKa = 7.65PNISRR6 pKa = 11.84IQALCRR12 pKa = 11.84MMMGLTLLAWAAARR26 pKa = 11.84FTRR29 pKa = 11.84RR30 pKa = 11.84PYY32 pKa = 10.85CPFSFIMALFGAQKK46 pKa = 8.89TAEE49 pKa = 4.13GTLRR53 pKa = 11.84FCPLVALRR61 pKa = 11.84KK62 pKa = 9.25RR63 pKa = 11.84CYY65 pKa = 10.33EE66 pKa = 3.84RR67 pKa = 11.84DD68 pKa = 3.31
Molecular weight: 7.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
755743 |
29 |
4605 |
292.8 |
32.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.285 ± 0.059 | 0.886 ± 0.018 |
5.187 ± 0.042 | 7.228 ± 0.061 |
4.223 ± 0.045 | 7.028 ± 0.064 |
2.513 ± 0.029 | 6.314 ± 0.043 |
5.659 ± 0.052 | 9.778 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.966 ± 0.027 | 3.324 ± 0.035 |
3.843 ± 0.051 | 4.011 ± 0.04 |
4.873 ± 0.045 | 5.429 ± 0.035 |
6.137 ± 0.065 | 7.891 ± 0.051 |
1.074 ± 0.021 | 3.35 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
