
Halogeometricum borinquense (strain ATCC 700274 / DSM 11551 / JCM 10706 / KCTC 4070 / PR3)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Halogeometricum; Halogeometricum borinquense
Average proteome isoelectric point is 4.99
Get precalculated fractions of proteins
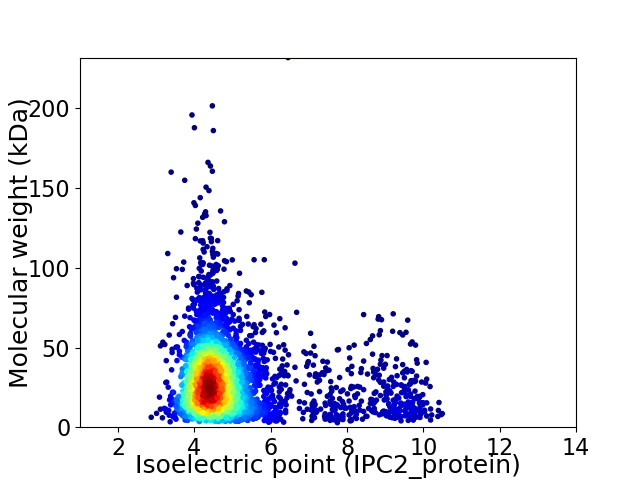
Virtual 2D-PAGE plot for 3894 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E4NLU9|E4NLU9_HALBP V-type ATP synthase subunit C OS=Halogeometricum borinquense (strain ATCC 700274 / DSM 11551 / JCM 10706 / KCTC 4070 / PR3) OX=469382 GN=atpC PE=3 SV=1
MM1 pKa = 7.6GLTEE5 pKa = 4.14TLDD8 pKa = 4.08DD9 pKa = 3.98YY10 pKa = 11.87FDD12 pKa = 3.91VQEE15 pKa = 4.88HH16 pKa = 6.16GSSVGTEE23 pKa = 3.53IVAGVTTFLTMSYY36 pKa = 9.62IVVVNPSILTDD47 pKa = 3.31QPFIEE52 pKa = 5.03GVDD55 pKa = 4.57GIAIAGFSPAEE66 pKa = 4.09VQSMLAVVTIVAAAVATFVMAIYY89 pKa = 10.71ANRR92 pKa = 11.84PFGQAPGLGLNAFFAFTVVGALGVPWQTALAAVVAEE128 pKa = 5.02GILFILLTAVGARR141 pKa = 11.84EE142 pKa = 3.72YY143 pKa = 10.82VIRR146 pKa = 11.84LFPEE150 pKa = 4.02PVKK153 pKa = 10.47FAVGTGIGLFLTIIGLQAMGLVVNDD178 pKa = 3.64SATLITLGPVASDD191 pKa = 3.35PVAILSVVGLFFTFALYY208 pKa = 10.75ARR210 pKa = 11.84GIPGSIIIGILGTAFLGWVLTVAGIVTPDD239 pKa = 3.12AGLVADD245 pKa = 5.07ASSMTLSLPVLGTFDD260 pKa = 3.73VLVPAAGASATYY272 pKa = 10.71NVTPLVGAFVGGLGNVEE289 pKa = 4.2AFSFALIVFTFFFVDD304 pKa = 4.8FFDD307 pKa = 3.79TAGTLVGVSQIAGFLDD323 pKa = 4.42DD324 pKa = 6.14DD325 pKa = 5.08GNLPDD330 pKa = 4.08IDD332 pKa = 4.47KK333 pKa = 10.7PLMADD338 pKa = 4.29AIGTTVGGMIGTSTVTTYY356 pKa = 10.82VEE358 pKa = 4.2SASGVEE364 pKa = 3.93EE365 pKa = 4.24GGRR368 pKa = 11.84TGLTALTVSVLFLVSLAFVPVAAAIPQYY396 pKa = 10.46ASHH399 pKa = 7.01IALVVIGVVMLGNVIDD415 pKa = 4.68IDD417 pKa = 3.57WSDD420 pKa = 3.64VTNTIPAGMTILVMPFTYY438 pKa = 10.44SIAYY442 pKa = 8.97GIAAGIVSYY451 pKa = 10.36PLVKK455 pKa = 9.66TAGGEE460 pKa = 4.01FDD462 pKa = 4.86DD463 pKa = 4.92VRR465 pKa = 11.84PGHH468 pKa = 6.59WALALAFVVYY478 pKa = 8.68FFVRR482 pKa = 11.84TSGIIQAQVV491 pKa = 2.47
MM1 pKa = 7.6GLTEE5 pKa = 4.14TLDD8 pKa = 4.08DD9 pKa = 3.98YY10 pKa = 11.87FDD12 pKa = 3.91VQEE15 pKa = 4.88HH16 pKa = 6.16GSSVGTEE23 pKa = 3.53IVAGVTTFLTMSYY36 pKa = 9.62IVVVNPSILTDD47 pKa = 3.31QPFIEE52 pKa = 5.03GVDD55 pKa = 4.57GIAIAGFSPAEE66 pKa = 4.09VQSMLAVVTIVAAAVATFVMAIYY89 pKa = 10.71ANRR92 pKa = 11.84PFGQAPGLGLNAFFAFTVVGALGVPWQTALAAVVAEE128 pKa = 5.02GILFILLTAVGARR141 pKa = 11.84EE142 pKa = 3.72YY143 pKa = 10.82VIRR146 pKa = 11.84LFPEE150 pKa = 4.02PVKK153 pKa = 10.47FAVGTGIGLFLTIIGLQAMGLVVNDD178 pKa = 3.64SATLITLGPVASDD191 pKa = 3.35PVAILSVVGLFFTFALYY208 pKa = 10.75ARR210 pKa = 11.84GIPGSIIIGILGTAFLGWVLTVAGIVTPDD239 pKa = 3.12AGLVADD245 pKa = 5.07ASSMTLSLPVLGTFDD260 pKa = 3.73VLVPAAGASATYY272 pKa = 10.71NVTPLVGAFVGGLGNVEE289 pKa = 4.2AFSFALIVFTFFFVDD304 pKa = 4.8FFDD307 pKa = 3.79TAGTLVGVSQIAGFLDD323 pKa = 4.42DD324 pKa = 6.14DD325 pKa = 5.08GNLPDD330 pKa = 4.08IDD332 pKa = 4.47KK333 pKa = 10.7PLMADD338 pKa = 4.29AIGTTVGGMIGTSTVTTYY356 pKa = 10.82VEE358 pKa = 4.2SASGVEE364 pKa = 3.93EE365 pKa = 4.24GGRR368 pKa = 11.84TGLTALTVSVLFLVSLAFVPVAAAIPQYY396 pKa = 10.46ASHH399 pKa = 7.01IALVVIGVVMLGNVIDD415 pKa = 4.68IDD417 pKa = 3.57WSDD420 pKa = 3.64VTNTIPAGMTILVMPFTYY438 pKa = 10.44SIAYY442 pKa = 8.97GIAAGIVSYY451 pKa = 10.36PLVKK455 pKa = 9.66TAGGEE460 pKa = 4.01FDD462 pKa = 4.86DD463 pKa = 4.92VRR465 pKa = 11.84PGHH468 pKa = 6.59WALALAFVVYY478 pKa = 8.68FFVRR482 pKa = 11.84TSGIIQAQVV491 pKa = 2.47
Molecular weight: 50.54 kDa
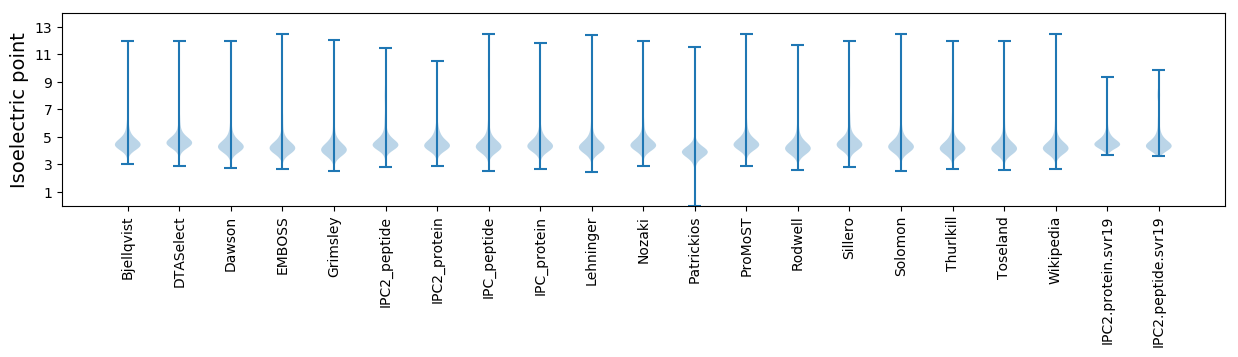
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E4NLG2|E4NLG2_HALBP Histidine--tRNA ligase OS=Halogeometricum borinquense (strain ATCC 700274 / DSM 11551 / JCM 10706 / KCTC 4070 / PR3) OX=469382 GN=hisS PE=3 SV=1
MM1 pKa = 7.26PTNDD5 pKa = 3.46AATKK9 pKa = 9.09RR10 pKa = 11.84TLEE13 pKa = 3.84KK14 pKa = 10.14QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NPARR27 pKa = 11.84ATSCRR32 pKa = 11.84KK33 pKa = 9.62CGYY36 pKa = 10.22KK37 pKa = 10.11NLRR40 pKa = 11.84PKK42 pKa = 10.56SKK44 pKa = 10.01EE45 pKa = 3.37RR46 pKa = 11.84RR47 pKa = 11.84AAA49 pKa = 3.42
MM1 pKa = 7.26PTNDD5 pKa = 3.46AATKK9 pKa = 9.09RR10 pKa = 11.84TLEE13 pKa = 3.84KK14 pKa = 10.14QICMRR19 pKa = 11.84CNARR23 pKa = 11.84NPARR27 pKa = 11.84ATSCRR32 pKa = 11.84KK33 pKa = 9.62CGYY36 pKa = 10.22KK37 pKa = 10.11NLRR40 pKa = 11.84PKK42 pKa = 10.56SKK44 pKa = 10.01EE45 pKa = 3.37RR46 pKa = 11.84RR47 pKa = 11.84AAA49 pKa = 3.42
Molecular weight: 5.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1130016 |
26 |
2192 |
290.2 |
31.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.204 ± 0.051 | 0.7 ± 0.013 |
8.155 ± 0.056 | 8.341 ± 0.056 |
3.475 ± 0.026 | 8.143 ± 0.039 |
2.026 ± 0.022 | 4.267 ± 0.031 |
2.255 ± 0.026 | 8.718 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.812 ± 0.018 | 2.714 ± 0.025 |
4.49 ± 0.028 | 2.676 ± 0.025 |
6.325 ± 0.041 | 6.167 ± 0.039 |
6.746 ± 0.042 | 8.929 ± 0.046 |
1.129 ± 0.015 | 2.726 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
