
Bat polyomavirus 6b
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus; Dobsonia moluccensis polyomavirus 2
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
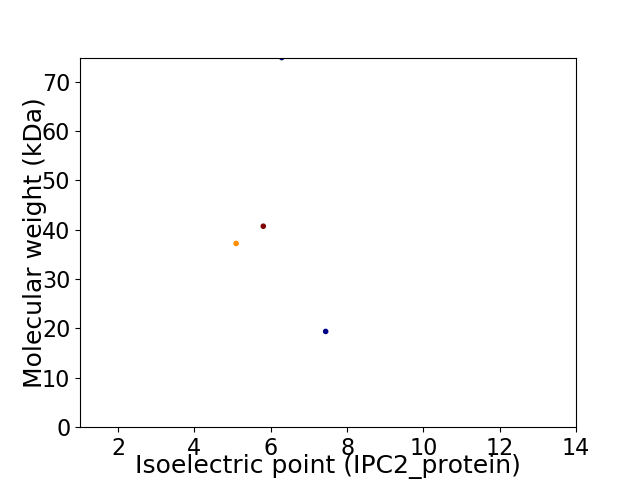
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
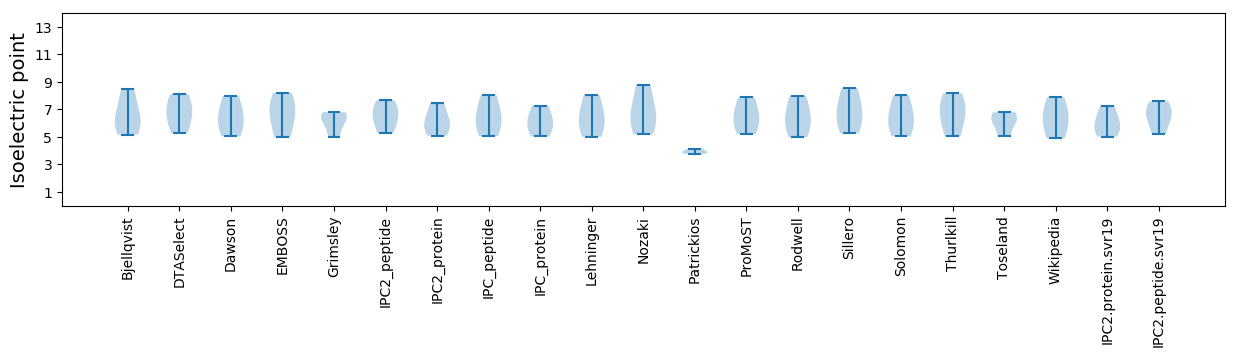
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5ZYR9|A0A0D5ZYR9_9POLY Large T antigen OS=Bat polyomavirus 6b OX=1623689 PE=4 SV=1
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.1VFEE13 pKa = 4.8LSAATGFTVDD23 pKa = 4.99SILTGEE29 pKa = 4.88AIVADD34 pKa = 4.73EE35 pKa = 4.7LLQAYY40 pKa = 5.77TTNLVALEE48 pKa = 4.71GFTQTEE54 pKa = 4.02ALLAAGFSPEE64 pKa = 3.7AAYY67 pKa = 10.99LLTSLAPNFPEE78 pKa = 5.08AFSLLAGAEE87 pKa = 4.41SVVHH91 pKa = 6.35GSLFIGAATSAALTPYY107 pKa = 10.05SYY109 pKa = 11.18DD110 pKa = 3.4YY111 pKa = 9.21ATPIADD117 pKa = 4.24LNQHH121 pKa = 6.35LMALQVWRR129 pKa = 11.84PEE131 pKa = 3.94DD132 pKa = 3.25WEE134 pKa = 5.37DD135 pKa = 3.25IYY137 pKa = 11.36FPGVLPFARR146 pKa = 11.84FVNYY150 pKa = 9.56IDD152 pKa = 3.88PANWASNLYY161 pKa = 9.65HH162 pKa = 7.6AIGRR166 pKa = 11.84YY167 pKa = 7.35FWEE170 pKa = 4.27SAQRR174 pKa = 11.84AGTRR178 pKa = 11.84LIEE181 pKa = 3.87QEE183 pKa = 4.14VRR185 pKa = 11.84HH186 pKa = 5.78VSTDD190 pKa = 3.12LAQRR194 pKa = 11.84TVTSIAEE201 pKa = 3.97TLSYY205 pKa = 10.91YY206 pKa = 10.62FEE208 pKa = 4.03NARR211 pKa = 11.84WAVSHH216 pKa = 6.72LSSNIYY222 pKa = 10.59GGLQQYY228 pKa = 10.22YY229 pKa = 10.38SEE231 pKa = 4.96LPPLRR236 pKa = 11.84PHH238 pKa = 6.27QVRR241 pKa = 11.84ALHH244 pKa = 6.22KK245 pKa = 10.28RR246 pKa = 11.84LGEE249 pKa = 4.35KK250 pKa = 10.17IPDD253 pKa = 3.66RR254 pKa = 11.84FNLEE258 pKa = 3.96SSKK261 pKa = 11.26GSAQYY266 pKa = 10.39VDD268 pKa = 5.36KK269 pKa = 11.08YY270 pKa = 10.81DD271 pKa = 3.62SPGGARR277 pKa = 11.84QRR279 pKa = 11.84HH280 pKa = 4.9TPDD283 pKa = 2.34WMLPLILGLYY293 pKa = 10.54GDD295 pKa = 5.2ILPSWEE301 pKa = 4.11TTLEE305 pKa = 3.97EE306 pKa = 5.13LEE308 pKa = 4.51AEE310 pKa = 4.08EE311 pKa = 6.19DD312 pKa = 4.16GPQKK316 pKa = 10.48KK317 pKa = 9.57KK318 pKa = 10.26PRR320 pKa = 11.84SEE322 pKa = 3.59THH324 pKa = 4.84RR325 pKa = 11.84RR326 pKa = 11.84RR327 pKa = 11.84RR328 pKa = 11.84KK329 pKa = 9.04SQSSAA334 pKa = 2.93
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.1VFEE13 pKa = 4.8LSAATGFTVDD23 pKa = 4.99SILTGEE29 pKa = 4.88AIVADD34 pKa = 4.73EE35 pKa = 4.7LLQAYY40 pKa = 5.77TTNLVALEE48 pKa = 4.71GFTQTEE54 pKa = 4.02ALLAAGFSPEE64 pKa = 3.7AAYY67 pKa = 10.99LLTSLAPNFPEE78 pKa = 5.08AFSLLAGAEE87 pKa = 4.41SVVHH91 pKa = 6.35GSLFIGAATSAALTPYY107 pKa = 10.05SYY109 pKa = 11.18DD110 pKa = 3.4YY111 pKa = 9.21ATPIADD117 pKa = 4.24LNQHH121 pKa = 6.35LMALQVWRR129 pKa = 11.84PEE131 pKa = 3.94DD132 pKa = 3.25WEE134 pKa = 5.37DD135 pKa = 3.25IYY137 pKa = 11.36FPGVLPFARR146 pKa = 11.84FVNYY150 pKa = 9.56IDD152 pKa = 3.88PANWASNLYY161 pKa = 9.65HH162 pKa = 7.6AIGRR166 pKa = 11.84YY167 pKa = 7.35FWEE170 pKa = 4.27SAQRR174 pKa = 11.84AGTRR178 pKa = 11.84LIEE181 pKa = 3.87QEE183 pKa = 4.14VRR185 pKa = 11.84HH186 pKa = 5.78VSTDD190 pKa = 3.12LAQRR194 pKa = 11.84TVTSIAEE201 pKa = 3.97TLSYY205 pKa = 10.91YY206 pKa = 10.62FEE208 pKa = 4.03NARR211 pKa = 11.84WAVSHH216 pKa = 6.72LSSNIYY222 pKa = 10.59GGLQQYY228 pKa = 10.22YY229 pKa = 10.38SEE231 pKa = 4.96LPPLRR236 pKa = 11.84PHH238 pKa = 6.27QVRR241 pKa = 11.84ALHH244 pKa = 6.22KK245 pKa = 10.28RR246 pKa = 11.84LGEE249 pKa = 4.35KK250 pKa = 10.17IPDD253 pKa = 3.66RR254 pKa = 11.84FNLEE258 pKa = 3.96SSKK261 pKa = 11.26GSAQYY266 pKa = 10.39VDD268 pKa = 5.36KK269 pKa = 11.08YY270 pKa = 10.81DD271 pKa = 3.62SPGGARR277 pKa = 11.84QRR279 pKa = 11.84HH280 pKa = 4.9TPDD283 pKa = 2.34WMLPLILGLYY293 pKa = 10.54GDD295 pKa = 5.2ILPSWEE301 pKa = 4.11TTLEE305 pKa = 3.97EE306 pKa = 5.13LEE308 pKa = 4.51AEE310 pKa = 4.08EE311 pKa = 6.19DD312 pKa = 4.16GPQKK316 pKa = 10.48KK317 pKa = 9.57KK318 pKa = 10.26PRR320 pKa = 11.84SEE322 pKa = 3.59THH324 pKa = 4.84RR325 pKa = 11.84RR326 pKa = 11.84RR327 pKa = 11.84RR328 pKa = 11.84KK329 pKa = 9.04SQSSAA334 pKa = 2.93
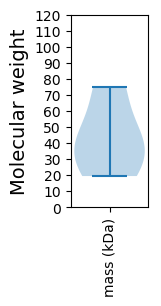
Molecular weight: 37.2 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5ZZ45|A0A0D5ZZ45_9POLY Small t antigen OS=Bat polyomavirus 6b OX=1623689 PE=4 SV=1
MM1 pKa = 7.69DD2 pKa = 4.33TGLSRR7 pKa = 11.84EE8 pKa = 3.95EE9 pKa = 4.02SQRR12 pKa = 11.84LLEE15 pKa = 4.72LLQLDD20 pKa = 4.16PEE22 pKa = 5.23HH23 pKa = 6.43YY24 pKa = 11.09GNWQLMRR31 pKa = 11.84KK32 pKa = 9.27SFLRR36 pKa = 11.84MCKK39 pKa = 9.6IMHH42 pKa = 7.06PDD44 pKa = 2.89KK45 pKa = 11.26GGNPEE50 pKa = 3.9AAKK53 pKa = 10.54EE54 pKa = 4.06LITLYY59 pKa = 10.81KK60 pKa = 10.42KK61 pKa = 10.91LEE63 pKa = 4.14NNISSLNPEE72 pKa = 3.94EE73 pKa = 5.31CFTTSQVEE81 pKa = 3.91KK82 pKa = 11.06SNFFLYY88 pKa = 10.25IKK90 pKa = 9.97DD91 pKa = 3.3WKK93 pKa = 8.56EE94 pKa = 4.02CNMGLKK100 pKa = 10.12PCVCIFCLTRR110 pKa = 11.84KK111 pKa = 7.77NHH113 pKa = 6.86KK114 pKa = 8.43EE115 pKa = 3.73RR116 pKa = 11.84KK117 pKa = 8.27NKK119 pKa = 9.31NLIWGKK125 pKa = 9.45CYY127 pKa = 10.57CFACYY132 pKa = 8.42CTWFGLEE139 pKa = 4.12WCWFTWLTWRR149 pKa = 11.84NIIAEE154 pKa = 4.42TPYY157 pKa = 10.65HH158 pKa = 6.47ALNLL162 pKa = 3.95
MM1 pKa = 7.69DD2 pKa = 4.33TGLSRR7 pKa = 11.84EE8 pKa = 3.95EE9 pKa = 4.02SQRR12 pKa = 11.84LLEE15 pKa = 4.72LLQLDD20 pKa = 4.16PEE22 pKa = 5.23HH23 pKa = 6.43YY24 pKa = 11.09GNWQLMRR31 pKa = 11.84KK32 pKa = 9.27SFLRR36 pKa = 11.84MCKK39 pKa = 9.6IMHH42 pKa = 7.06PDD44 pKa = 2.89KK45 pKa = 11.26GGNPEE50 pKa = 3.9AAKK53 pKa = 10.54EE54 pKa = 4.06LITLYY59 pKa = 10.81KK60 pKa = 10.42KK61 pKa = 10.91LEE63 pKa = 4.14NNISSLNPEE72 pKa = 3.94EE73 pKa = 5.31CFTTSQVEE81 pKa = 3.91KK82 pKa = 11.06SNFFLYY88 pKa = 10.25IKK90 pKa = 9.97DD91 pKa = 3.3WKK93 pKa = 8.56EE94 pKa = 4.02CNMGLKK100 pKa = 10.12PCVCIFCLTRR110 pKa = 11.84KK111 pKa = 7.77NHH113 pKa = 6.86KK114 pKa = 8.43EE115 pKa = 3.73RR116 pKa = 11.84KK117 pKa = 8.27NKK119 pKa = 9.31NLIWGKK125 pKa = 9.45CYY127 pKa = 10.57CFACYY132 pKa = 8.42CTWFGLEE139 pKa = 4.12WCWFTWLTWRR149 pKa = 11.84NIIAEE154 pKa = 4.42TPYY157 pKa = 10.65HH158 pKa = 6.47ALNLL162 pKa = 3.95
Molecular weight: 19.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1517 |
162 |
652 |
379.3 |
43.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.262 ± 1.518 | 2.373 ± 0.85 |
4.285 ± 0.358 | 7.449 ± 0.321 |
4.417 ± 0.382 | 5.735 ± 1.016 |
2.175 ± 0.259 | 4.944 ± 0.325 |
6.394 ± 1.242 | 10.481 ± 0.913 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.175 ± 0.333 | 5.339 ± 0.683 |
5.735 ± 0.409 | 4.548 ± 0.375 |
4.417 ± 0.36 | 6.658 ± 0.452 |
5.603 ± 0.128 | 5.405 ± 1.039 |
1.582 ± 0.602 | 4.021 ± 0.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
