
Paraoerskovia marina
Taxonomy:
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
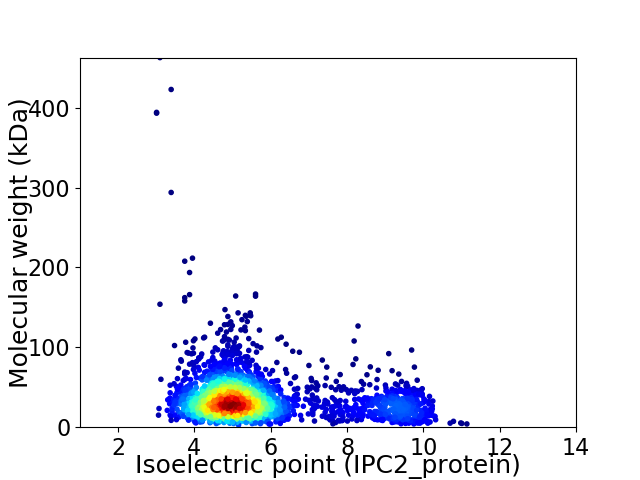
Virtual 2D-PAGE plot for 2637 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1NHU9|A0A1H1NHU9_9CELL ATP-dependent helicase HrpA OS=Paraoerskovia marina OX=545619 GN=SAMN04489860_0540 PE=4 SV=1
MM1 pKa = 7.21HH2 pKa = 7.98RR3 pKa = 11.84SPTILPDD10 pKa = 3.64AQPPRR15 pKa = 11.84ARR17 pKa = 11.84HH18 pKa = 4.81GVALVAATALLALAGTTTLAAVTLDD43 pKa = 3.99ADD45 pKa = 3.91PAAAAGTCEE54 pKa = 5.29ADD56 pKa = 3.79FDD58 pKa = 6.07VNNQWNGGFGTNVSVTSPGDD78 pKa = 3.41PVTSWEE84 pKa = 4.4VTWEE88 pKa = 4.04FSGDD92 pKa = 3.65QQVTQLWNGTVSQSGNTVHH111 pKa = 6.63VASLPWNGAVGTGGTLSFGFNGSGDD136 pKa = 3.64GSTATPTAVTLNGAACSTGTDD157 pKa = 3.69PTPTPTTTTPTPTPTEE173 pKa = 4.16TTPTPTPTEE182 pKa = 4.23TTDD185 pKa = 3.23PPAADD190 pKa = 3.33GDD192 pKa = 4.58LYY194 pKa = 11.64VDD196 pKa = 4.45TGNQAYY202 pKa = 9.43AAWSAASGDD211 pKa = 4.13DD212 pKa = 3.33KK213 pKa = 11.9ALLAKK218 pKa = 10.24IALTPQAYY226 pKa = 7.69WVGNWDD232 pKa = 4.34DD233 pKa = 4.36PEE235 pKa = 4.42HH236 pKa = 6.28AQAEE240 pKa = 4.57VQDD243 pKa = 3.95YY244 pKa = 9.46TGRR247 pKa = 11.84AVAAGQVPQLVIYY260 pKa = 8.6AIPGRR265 pKa = 11.84DD266 pKa = 3.3CGSYY270 pKa = 10.22SGGGVAEE277 pKa = 4.34SEE279 pKa = 4.2YY280 pKa = 11.37ADD282 pKa = 4.59WIDD285 pKa = 3.39TMAAGIQGNPLVVLEE300 pKa = 4.69PDD302 pKa = 3.57ALAQLGDD309 pKa = 4.21CDD311 pKa = 4.1GQGDD315 pKa = 4.01RR316 pKa = 11.84VGYY319 pKa = 9.38LAYY322 pKa = 10.05AASSLTDD329 pKa = 3.48AGAHH333 pKa = 5.21VYY335 pKa = 10.11IDD337 pKa = 4.21AGHH340 pKa = 6.29SGWLTPATAADD351 pKa = 4.0RR352 pKa = 11.84LLQVGLDD359 pKa = 3.79DD360 pKa = 4.65AVGFAVNTSNYY371 pKa = 7.05QTTADD376 pKa = 3.56SQAWAEE382 pKa = 4.09QVSALTGGAGYY393 pKa = 10.87VIDD396 pKa = 4.08TSRR399 pKa = 11.84NGNGSNGQWCNPSGRR414 pKa = 11.84ALGDD418 pKa = 3.43RR419 pKa = 11.84PSLVDD424 pKa = 5.11DD425 pKa = 4.11STNLDD430 pKa = 3.09ALLWVKK436 pKa = 10.69LPGEE440 pKa = 4.21SDD442 pKa = 3.59GTCNGGPSAGAWWQSMALEE461 pKa = 4.5LARR464 pKa = 11.84NASWW468 pKa = 3.09
MM1 pKa = 7.21HH2 pKa = 7.98RR3 pKa = 11.84SPTILPDD10 pKa = 3.64AQPPRR15 pKa = 11.84ARR17 pKa = 11.84HH18 pKa = 4.81GVALVAATALLALAGTTTLAAVTLDD43 pKa = 3.99ADD45 pKa = 3.91PAAAAGTCEE54 pKa = 5.29ADD56 pKa = 3.79FDD58 pKa = 6.07VNNQWNGGFGTNVSVTSPGDD78 pKa = 3.41PVTSWEE84 pKa = 4.4VTWEE88 pKa = 4.04FSGDD92 pKa = 3.65QQVTQLWNGTVSQSGNTVHH111 pKa = 6.63VASLPWNGAVGTGGTLSFGFNGSGDD136 pKa = 3.64GSTATPTAVTLNGAACSTGTDD157 pKa = 3.69PTPTPTTTTPTPTPTEE173 pKa = 4.16TTPTPTPTEE182 pKa = 4.23TTDD185 pKa = 3.23PPAADD190 pKa = 3.33GDD192 pKa = 4.58LYY194 pKa = 11.64VDD196 pKa = 4.45TGNQAYY202 pKa = 9.43AAWSAASGDD211 pKa = 4.13DD212 pKa = 3.33KK213 pKa = 11.9ALLAKK218 pKa = 10.24IALTPQAYY226 pKa = 7.69WVGNWDD232 pKa = 4.34DD233 pKa = 4.36PEE235 pKa = 4.42HH236 pKa = 6.28AQAEE240 pKa = 4.57VQDD243 pKa = 3.95YY244 pKa = 9.46TGRR247 pKa = 11.84AVAAGQVPQLVIYY260 pKa = 8.6AIPGRR265 pKa = 11.84DD266 pKa = 3.3CGSYY270 pKa = 10.22SGGGVAEE277 pKa = 4.34SEE279 pKa = 4.2YY280 pKa = 11.37ADD282 pKa = 4.59WIDD285 pKa = 3.39TMAAGIQGNPLVVLEE300 pKa = 4.69PDD302 pKa = 3.57ALAQLGDD309 pKa = 4.21CDD311 pKa = 4.1GQGDD315 pKa = 4.01RR316 pKa = 11.84VGYY319 pKa = 9.38LAYY322 pKa = 10.05AASSLTDD329 pKa = 3.48AGAHH333 pKa = 5.21VYY335 pKa = 10.11IDD337 pKa = 4.21AGHH340 pKa = 6.29SGWLTPATAADD351 pKa = 4.0RR352 pKa = 11.84LLQVGLDD359 pKa = 3.79DD360 pKa = 4.65AVGFAVNTSNYY371 pKa = 7.05QTTADD376 pKa = 3.56SQAWAEE382 pKa = 4.09QVSALTGGAGYY393 pKa = 10.87VIDD396 pKa = 4.08TSRR399 pKa = 11.84NGNGSNGQWCNPSGRR414 pKa = 11.84ALGDD418 pKa = 3.43RR419 pKa = 11.84PSLVDD424 pKa = 5.11DD425 pKa = 4.11STNLDD430 pKa = 3.09ALLWVKK436 pKa = 10.69LPGEE440 pKa = 4.21SDD442 pKa = 3.59GTCNGGPSAGAWWQSMALEE461 pKa = 4.5LARR464 pKa = 11.84NASWW468 pKa = 3.09
Molecular weight: 47.86 kDa
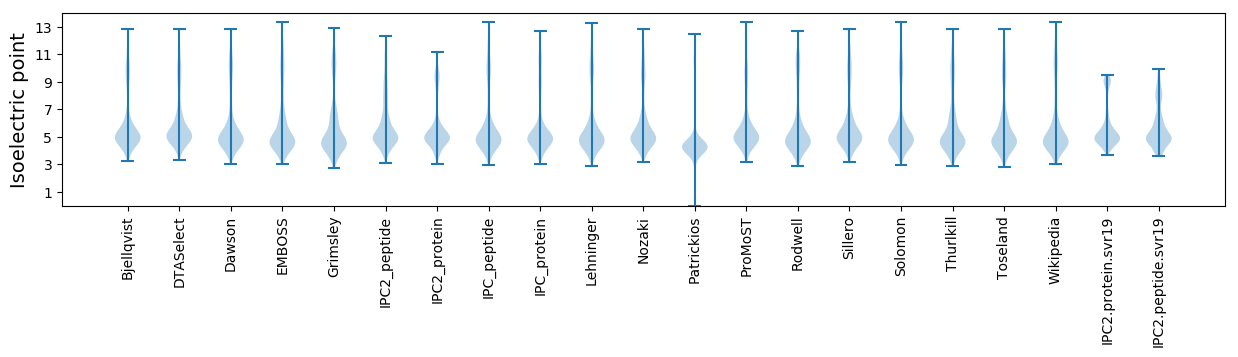
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1VMW4|A0A1H1VMW4_9CELL DNA-binding transcriptional regulator LacI/PurR family OS=Paraoerskovia marina OX=545619 GN=SAMN04489860_2553 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
905901 |
25 |
4501 |
343.5 |
36.57 |
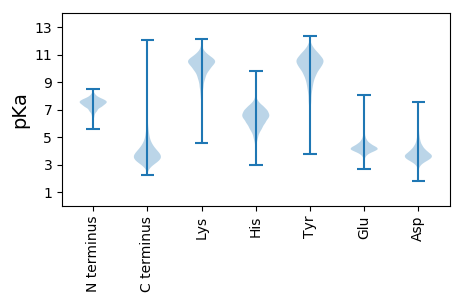
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.289 ± 0.079 | 0.612 ± 0.012 |
6.976 ± 0.056 | 5.933 ± 0.043 |
2.666 ± 0.027 | 9.189 ± 0.042 |
1.986 ± 0.026 | 3.486 ± 0.037 |
1.635 ± 0.036 | 9.706 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.697 ± 0.022 | 1.743 ± 0.028 |
5.619 ± 0.041 | 2.56 ± 0.025 |
7.293 ± 0.073 | 5.572 ± 0.033 |
6.59 ± 0.06 | 10.025 ± 0.054 |
1.503 ± 0.022 | 1.917 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
