
Botrytis virus X (isolate Botrytis cinerea/New Zealand/Howitt/2006) (BOTV-X)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Alphaflexiviridae; Botrexvirus; Botrytis virus X
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
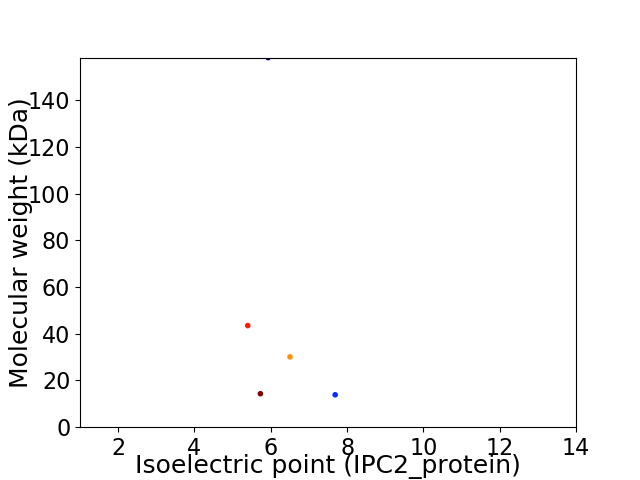
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6YNQ6|ORF2_BOTVX Uncharacterized ORF2 protein OS=Botrytis virus X (isolate Botrytis cinerea/New Zealand/Howitt/2006) OX=686947 GN=ORF2 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 6.13PNLDD6 pKa = 3.52QDD8 pKa = 4.01TLPTHH13 pKa = 6.28EE14 pKa = 5.49EE15 pKa = 3.43IDD17 pKa = 3.64NDD19 pKa = 3.22VDD21 pKa = 3.96SAEE24 pKa = 4.44EE25 pKa = 4.17EE26 pKa = 4.47PPEE29 pKa = 4.71PPLLPDD35 pKa = 6.43DD36 pKa = 4.92IDD38 pKa = 6.46DD39 pKa = 5.03DD40 pKa = 4.91DD41 pKa = 4.31SHH43 pKa = 7.56GSRR46 pKa = 11.84TRR48 pKa = 11.84RR49 pKa = 11.84QVKK52 pKa = 9.46PPPEE56 pKa = 3.96LLRR59 pKa = 11.84AVGACLISGHH69 pKa = 6.0YY70 pKa = 10.5DD71 pKa = 2.82GGNYY75 pKa = 9.11FRR77 pKa = 11.84WQQSIAALYY86 pKa = 10.27AKK88 pKa = 10.08AGYY91 pKa = 10.43AGDD94 pKa = 3.63IRR96 pKa = 11.84FHH98 pKa = 5.58QAAIQEE104 pKa = 4.39YY105 pKa = 9.98ALDD108 pKa = 4.12PVLPAPRR115 pKa = 11.84VSYY118 pKa = 11.09DD119 pKa = 3.62LLVAHH124 pKa = 7.34AGLRR128 pKa = 11.84YY129 pKa = 8.98QALLNEE135 pKa = 4.4QLRR138 pKa = 11.84TGKK141 pKa = 8.24TPPADD146 pKa = 3.73EE147 pKa = 4.38ALKK150 pKa = 10.9DD151 pKa = 3.92AVRR154 pKa = 11.84KK155 pKa = 8.83AAQAAYY161 pKa = 10.36DD162 pKa = 3.92NAVKK166 pKa = 9.95TGDD169 pKa = 3.88YY170 pKa = 10.13TPLIDD175 pKa = 4.17IAFKK179 pKa = 10.78GVDD182 pKa = 3.64INKK185 pKa = 9.22HH186 pKa = 5.38ASDD189 pKa = 3.78VAQLAKK195 pKa = 10.25MSVTMDD201 pKa = 3.05GTHH204 pKa = 6.45IKK206 pKa = 8.88FTAGEE211 pKa = 4.12MPKK214 pKa = 10.87DD215 pKa = 3.53KK216 pKa = 11.04VITSNSMASPNTVMNILNLITTSANVTAVTCGIEE250 pKa = 3.86FAIACAHH257 pKa = 5.87QGSSRR262 pKa = 11.84YY263 pKa = 7.37TRR265 pKa = 11.84HH266 pKa = 6.2TGTSTGGSTFEE277 pKa = 5.42LIAAHH282 pKa = 5.85VKK284 pKa = 7.25EE285 pKa = 4.64HH286 pKa = 5.26CTIRR290 pKa = 11.84QFCSYY295 pKa = 9.46FAKK298 pKa = 10.67VVWNHH303 pKa = 6.35LLTHH307 pKa = 6.5ATPPVNWAKK316 pKa = 10.73HH317 pKa = 4.94GFTLDD322 pKa = 2.93SRR324 pKa = 11.84YY325 pKa = 10.45AAFDD329 pKa = 3.5FFDD332 pKa = 3.86AVTNAAALPPKK343 pKa = 10.12NGLIRR348 pKa = 11.84APTSEE353 pKa = 4.91EE354 pKa = 3.46IRR356 pKa = 11.84AHH358 pKa = 6.02NLNAHH363 pKa = 6.37LLINASRR370 pKa = 11.84QDD372 pKa = 3.76DD373 pKa = 4.04QVSSSAQYY381 pKa = 9.0TAAIAQAGGFKK392 pKa = 10.3RR393 pKa = 11.84PQIGWGEE400 pKa = 3.9
MM1 pKa = 8.1DD2 pKa = 6.13PNLDD6 pKa = 3.52QDD8 pKa = 4.01TLPTHH13 pKa = 6.28EE14 pKa = 5.49EE15 pKa = 3.43IDD17 pKa = 3.64NDD19 pKa = 3.22VDD21 pKa = 3.96SAEE24 pKa = 4.44EE25 pKa = 4.17EE26 pKa = 4.47PPEE29 pKa = 4.71PPLLPDD35 pKa = 6.43DD36 pKa = 4.92IDD38 pKa = 6.46DD39 pKa = 5.03DD40 pKa = 4.91DD41 pKa = 4.31SHH43 pKa = 7.56GSRR46 pKa = 11.84TRR48 pKa = 11.84RR49 pKa = 11.84QVKK52 pKa = 9.46PPPEE56 pKa = 3.96LLRR59 pKa = 11.84AVGACLISGHH69 pKa = 6.0YY70 pKa = 10.5DD71 pKa = 2.82GGNYY75 pKa = 9.11FRR77 pKa = 11.84WQQSIAALYY86 pKa = 10.27AKK88 pKa = 10.08AGYY91 pKa = 10.43AGDD94 pKa = 3.63IRR96 pKa = 11.84FHH98 pKa = 5.58QAAIQEE104 pKa = 4.39YY105 pKa = 9.98ALDD108 pKa = 4.12PVLPAPRR115 pKa = 11.84VSYY118 pKa = 11.09DD119 pKa = 3.62LLVAHH124 pKa = 7.34AGLRR128 pKa = 11.84YY129 pKa = 8.98QALLNEE135 pKa = 4.4QLRR138 pKa = 11.84TGKK141 pKa = 8.24TPPADD146 pKa = 3.73EE147 pKa = 4.38ALKK150 pKa = 10.9DD151 pKa = 3.92AVRR154 pKa = 11.84KK155 pKa = 8.83AAQAAYY161 pKa = 10.36DD162 pKa = 3.92NAVKK166 pKa = 9.95TGDD169 pKa = 3.88YY170 pKa = 10.13TPLIDD175 pKa = 4.17IAFKK179 pKa = 10.78GVDD182 pKa = 3.64INKK185 pKa = 9.22HH186 pKa = 5.38ASDD189 pKa = 3.78VAQLAKK195 pKa = 10.25MSVTMDD201 pKa = 3.05GTHH204 pKa = 6.45IKK206 pKa = 8.88FTAGEE211 pKa = 4.12MPKK214 pKa = 10.87DD215 pKa = 3.53KK216 pKa = 11.04VITSNSMASPNTVMNILNLITTSANVTAVTCGIEE250 pKa = 3.86FAIACAHH257 pKa = 5.87QGSSRR262 pKa = 11.84YY263 pKa = 7.37TRR265 pKa = 11.84HH266 pKa = 6.2TGTSTGGSTFEE277 pKa = 5.42LIAAHH282 pKa = 5.85VKK284 pKa = 7.25EE285 pKa = 4.64HH286 pKa = 5.26CTIRR290 pKa = 11.84QFCSYY295 pKa = 9.46FAKK298 pKa = 10.67VVWNHH303 pKa = 6.35LLTHH307 pKa = 6.5ATPPVNWAKK316 pKa = 10.73HH317 pKa = 4.94GFTLDD322 pKa = 2.93SRR324 pKa = 11.84YY325 pKa = 10.45AAFDD329 pKa = 3.5FFDD332 pKa = 3.86AVTNAAALPPKK343 pKa = 10.12NGLIRR348 pKa = 11.84APTSEE353 pKa = 4.91EE354 pKa = 3.46IRR356 pKa = 11.84AHH358 pKa = 6.02NLNAHH363 pKa = 6.37LLINASRR370 pKa = 11.84QDD372 pKa = 3.76DD373 pKa = 4.04QVSSSAQYY381 pKa = 9.0TAAIAQAGGFKK392 pKa = 10.3RR393 pKa = 11.84PQIGWGEE400 pKa = 3.9
Molecular weight: 43.5 kDa
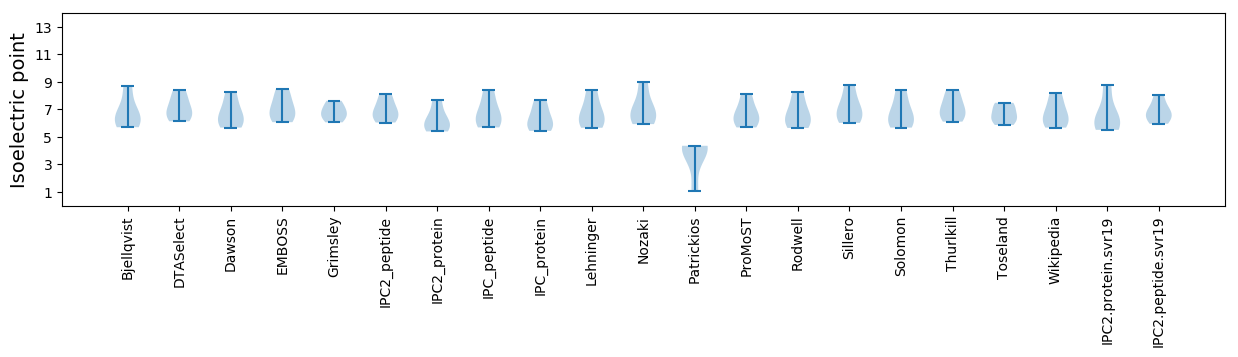
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q6YNQ5|CAPSD_BOTVX Capsid protein OS=Botrytis virus X (isolate Botrytis cinerea/New Zealand/Howitt/2006) OX=686947 GN=ORF3 PE=3 SV=1
MM1 pKa = 7.16PTYY4 pKa = 11.16SSMLHH9 pKa = 5.91AKK11 pKa = 9.06MIKK14 pKa = 10.1SPAALNTPQPSPKK27 pKa = 9.52QAVSNAHH34 pKa = 6.49RR35 pKa = 11.84LDD37 pKa = 4.14GGSSYY42 pKa = 11.09VVSLSEE48 pKa = 4.32ISSCPTSTTNSLFAAATPSTLLLSALYY75 pKa = 7.47CTPPSQLRR83 pKa = 11.84MKK85 pKa = 10.51SSCTLTLFIPPLNTAASPTLPDD107 pKa = 3.93TSCHH111 pKa = 6.07GEE113 pKa = 3.9WNTTTRR119 pKa = 11.84PCLAASTALMYY130 pKa = 10.98SLL132 pKa = 5.14
MM1 pKa = 7.16PTYY4 pKa = 11.16SSMLHH9 pKa = 5.91AKK11 pKa = 9.06MIKK14 pKa = 10.1SPAALNTPQPSPKK27 pKa = 9.52QAVSNAHH34 pKa = 6.49RR35 pKa = 11.84LDD37 pKa = 4.14GGSSYY42 pKa = 11.09VVSLSEE48 pKa = 4.32ISSCPTSTTNSLFAAATPSTLLLSALYY75 pKa = 7.47CTPPSQLRR83 pKa = 11.84MKK85 pKa = 10.51SSCTLTLFIPPLNTAASPTLPDD107 pKa = 3.93TSCHH111 pKa = 6.07GEE113 pKa = 3.9WNTTTRR119 pKa = 11.84PCLAASTALMYY130 pKa = 10.98SLL132 pKa = 5.14
Molecular weight: 13.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2351 |
128 |
1413 |
470.2 |
52.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.485 ± 1.041 | 1.148 ± 0.41 |
6.125 ± 0.791 | 4.466 ± 0.692 |
4.041 ± 0.459 | 4.381 ± 0.539 |
4.339 ± 0.502 | 4.892 ± 0.852 |
4.806 ± 0.812 | 10.038 ± 0.795 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.829 ± 0.43 | 4.083 ± 0.233 |
8.507 ± 1.763 | 3.488 ± 0.572 |
4.424 ± 0.429 | 6.465 ± 1.43 |
8.762 ± 0.564 | 5.019 ± 0.451 |
0.808 ± 0.046 | 2.892 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
