
Gibberella nygamai (Bean root rot disease fungus) (Fusarium nygamai)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta;
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
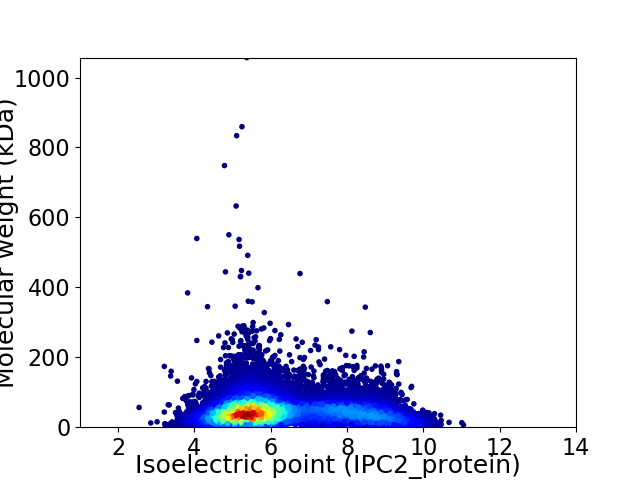
Virtual 2D-PAGE plot for 16018 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K0WJ50|A0A2K0WJ50_GIBNY Uncharacterized protein OS=Gibberella nygamai OX=42673 GN=FNYG_04497 PE=3 SV=1
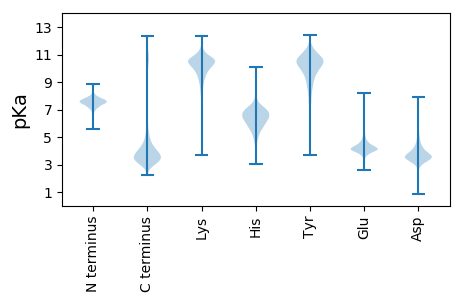
MM1 pKa = 7.38KK2 pKa = 10.2NAAALLTLLPLVAAHH17 pKa = 6.66GFVKK21 pKa = 10.42SPPPRR26 pKa = 11.84KK27 pKa = 9.37PGNAFKK33 pKa = 10.35AACGEE38 pKa = 4.09QPFYY42 pKa = 10.66QQSADD47 pKa = 3.16INGNVQGIKK56 pKa = 10.15QVVGSNFDD64 pKa = 3.66AKK66 pKa = 10.7EE67 pKa = 4.01CNLWLCKK74 pKa = 10.37GFQFDD79 pKa = 4.17DD80 pKa = 3.81NKK82 pKa = 11.6DD83 pKa = 3.44NVQSYY88 pKa = 11.28KK89 pKa = 10.68LGEE92 pKa = 4.64KK93 pKa = 9.51IDD95 pKa = 3.82FDD97 pKa = 4.79VNIAAPHH104 pKa = 5.53TGYY107 pKa = 11.49ANVSVVKK114 pKa = 10.03TSTDD118 pKa = 2.94KK119 pKa = 11.05MIGEE123 pKa = 4.16PLIEE127 pKa = 4.31FEE129 pKa = 5.05NYY131 pKa = 9.16AANAGLNPNNTAFSVTLPEE150 pKa = 4.25SLGGEE155 pKa = 4.23CTKK158 pKa = 11.08AGDD161 pKa = 5.53CVLQWFWDD169 pKa = 3.95APDD172 pKa = 3.63IDD174 pKa = 4.14QTYY177 pKa = 8.91EE178 pKa = 3.82SCVDD182 pKa = 3.9FVVGAGSGSGSGSGSGSTKK201 pKa = 9.87PSSAASAAPVATSAPAAEE219 pKa = 4.91KK220 pKa = 8.85PTATTLQAVAVTSSALGPVFEE241 pKa = 5.12SVTTAVPEE249 pKa = 4.29PTATTPDD256 pKa = 3.36AGDD259 pKa = 5.65DD260 pKa = 3.81EE261 pKa = 6.38DD262 pKa = 6.47CDD264 pKa = 4.51DD265 pKa = 5.14EE266 pKa = 5.32EE267 pKa = 5.61PVDD270 pKa = 4.04QGDD273 pKa = 6.0DD274 pKa = 3.64EE275 pKa = 6.24DD276 pKa = 6.45CDD278 pKa = 4.8DD279 pKa = 5.11EE280 pKa = 5.27EE281 pKa = 4.64PTPSAAAEE289 pKa = 4.11TGDD292 pKa = 6.21DD293 pKa = 3.96EE294 pKa = 6.58DD295 pKa = 5.64CDD297 pKa = 3.75EE298 pKa = 5.29DD299 pKa = 4.36EE300 pKa = 4.72EE301 pKa = 5.64PEE303 pKa = 4.18TGDD306 pKa = 5.32DD307 pKa = 5.47DD308 pKa = 4.67EE309 pKa = 5.36EE310 pKa = 5.72CPADD314 pKa = 5.98DD315 pKa = 4.24GDD317 pKa = 5.08EE318 pKa = 4.53YY319 pKa = 11.01DD320 pKa = 4.32TQPATPSNTAQGISAAANTAAVTKK344 pKa = 7.76TQNNAYY350 pKa = 8.16PVPKK354 pKa = 8.76PTGHH358 pKa = 6.35SSSDD362 pKa = 3.31KK363 pKa = 10.36TGSGSKK369 pKa = 10.67GSNNNNNAGSNYY381 pKa = 9.91NNYY384 pKa = 9.75GNSGSNNNNAGSNTIMTSYY403 pKa = 9.19VTVSAAEE410 pKa = 3.91THH412 pKa = 5.86YY413 pKa = 10.77VTVTADD419 pKa = 3.51APACTAA425 pKa = 4.06
MM1 pKa = 7.38KK2 pKa = 10.2NAAALLTLLPLVAAHH17 pKa = 6.66GFVKK21 pKa = 10.42SPPPRR26 pKa = 11.84KK27 pKa = 9.37PGNAFKK33 pKa = 10.35AACGEE38 pKa = 4.09QPFYY42 pKa = 10.66QQSADD47 pKa = 3.16INGNVQGIKK56 pKa = 10.15QVVGSNFDD64 pKa = 3.66AKK66 pKa = 10.7EE67 pKa = 4.01CNLWLCKK74 pKa = 10.37GFQFDD79 pKa = 4.17DD80 pKa = 3.81NKK82 pKa = 11.6DD83 pKa = 3.44NVQSYY88 pKa = 11.28KK89 pKa = 10.68LGEE92 pKa = 4.64KK93 pKa = 9.51IDD95 pKa = 3.82FDD97 pKa = 4.79VNIAAPHH104 pKa = 5.53TGYY107 pKa = 11.49ANVSVVKK114 pKa = 10.03TSTDD118 pKa = 2.94KK119 pKa = 11.05MIGEE123 pKa = 4.16PLIEE127 pKa = 4.31FEE129 pKa = 5.05NYY131 pKa = 9.16AANAGLNPNNTAFSVTLPEE150 pKa = 4.25SLGGEE155 pKa = 4.23CTKK158 pKa = 11.08AGDD161 pKa = 5.53CVLQWFWDD169 pKa = 3.95APDD172 pKa = 3.63IDD174 pKa = 4.14QTYY177 pKa = 8.91EE178 pKa = 3.82SCVDD182 pKa = 3.9FVVGAGSGSGSGSGSGSTKK201 pKa = 9.87PSSAASAAPVATSAPAAEE219 pKa = 4.91KK220 pKa = 8.85PTATTLQAVAVTSSALGPVFEE241 pKa = 5.12SVTTAVPEE249 pKa = 4.29PTATTPDD256 pKa = 3.36AGDD259 pKa = 5.65DD260 pKa = 3.81EE261 pKa = 6.38DD262 pKa = 6.47CDD264 pKa = 4.51DD265 pKa = 5.14EE266 pKa = 5.32EE267 pKa = 5.61PVDD270 pKa = 4.04QGDD273 pKa = 6.0DD274 pKa = 3.64EE275 pKa = 6.24DD276 pKa = 6.45CDD278 pKa = 4.8DD279 pKa = 5.11EE280 pKa = 5.27EE281 pKa = 4.64PTPSAAAEE289 pKa = 4.11TGDD292 pKa = 6.21DD293 pKa = 3.96EE294 pKa = 6.58DD295 pKa = 5.64CDD297 pKa = 3.75EE298 pKa = 5.29DD299 pKa = 4.36EE300 pKa = 4.72EE301 pKa = 5.64PEE303 pKa = 4.18TGDD306 pKa = 5.32DD307 pKa = 5.47DD308 pKa = 4.67EE309 pKa = 5.36EE310 pKa = 5.72CPADD314 pKa = 5.98DD315 pKa = 4.24GDD317 pKa = 5.08EE318 pKa = 4.53YY319 pKa = 11.01DD320 pKa = 4.32TQPATPSNTAQGISAAANTAAVTKK344 pKa = 7.76TQNNAYY350 pKa = 8.16PVPKK354 pKa = 8.76PTGHH358 pKa = 6.35SSSDD362 pKa = 3.31KK363 pKa = 10.36TGSGSKK369 pKa = 10.67GSNNNNNAGSNYY381 pKa = 9.91NNYY384 pKa = 9.75GNSGSNNNNAGSNTIMTSYY403 pKa = 9.19VTVSAAEE410 pKa = 3.91THH412 pKa = 5.86YY413 pKa = 10.77VTVTADD419 pKa = 3.51APACTAA425 pKa = 4.06
Molecular weight: 43.8 kDa
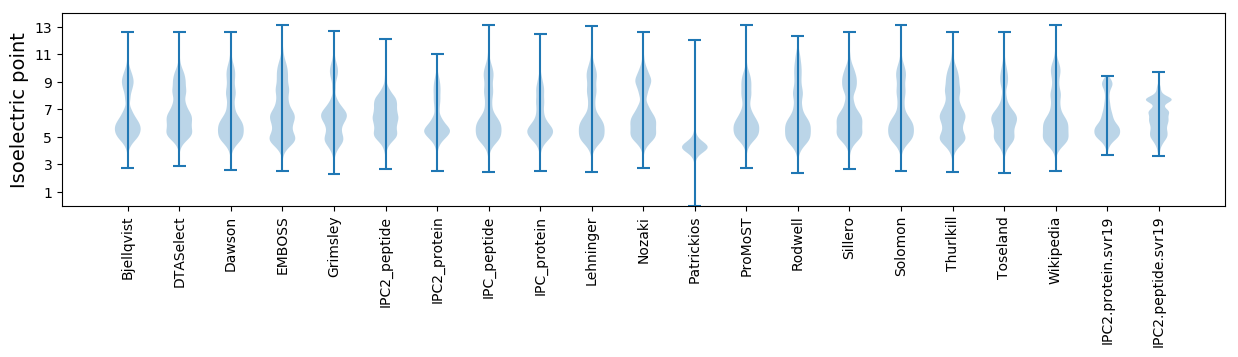
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K0WBY0|A0A2K0WBY0_GIBNY Uncharacterized protein OS=Gibberella nygamai OX=42673 GN=FNYG_06869 PE=4 SV=1
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APAHH26 pKa = 4.84SHH28 pKa = 5.33RR29 pKa = 11.84HH30 pKa = 4.03TTTTTTTTTKK40 pKa = 9.5PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGAGRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 6.46ATTTAPVHH64 pKa = 4.87HH65 pKa = 6.63HH66 pKa = 5.14QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.67GSLTRR89 pKa = 11.84QPGVKK94 pKa = 9.99AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
MM1 pKa = 7.86PLTRR5 pKa = 11.84THH7 pKa = 6.63RR8 pKa = 11.84HH9 pKa = 4.04TAPRR13 pKa = 11.84RR14 pKa = 11.84SIFSTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84APAHH26 pKa = 4.84SHH28 pKa = 5.33RR29 pKa = 11.84HH30 pKa = 4.03TTTTTTTTTKK40 pKa = 9.5PRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84GMFGGGAGRR53 pKa = 11.84RR54 pKa = 11.84THH56 pKa = 6.46ATTTAPVHH64 pKa = 4.87HH65 pKa = 6.63HH66 pKa = 5.14QRR68 pKa = 11.84RR69 pKa = 11.84PSMKK73 pKa = 10.02DD74 pKa = 2.95KK75 pKa = 11.41VSGALLKK82 pKa = 11.04LKK84 pKa = 10.67GSLTRR89 pKa = 11.84QPGVKK94 pKa = 9.99AAGTRR99 pKa = 11.84RR100 pKa = 11.84MRR102 pKa = 11.84GTDD105 pKa = 3.02GRR107 pKa = 11.84GARR110 pKa = 11.84HH111 pKa = 5.87HH112 pKa = 7.23RR113 pKa = 11.84YY114 pKa = 9.44
Molecular weight: 12.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7249913 |
30 |
9543 |
452.6 |
50.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.382 ± 0.018 | 1.264 ± 0.008 |
5.919 ± 0.013 | 6.276 ± 0.021 |
3.792 ± 0.012 | 6.817 ± 0.019 |
2.365 ± 0.008 | 5.137 ± 0.014 |
5.083 ± 0.019 | 8.814 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.241 ± 0.008 | 3.781 ± 0.01 |
5.843 ± 0.022 | 3.991 ± 0.015 |
5.81 ± 0.016 | 7.992 ± 0.02 |
5.984 ± 0.022 | 6.126 ± 0.014 |
1.56 ± 0.007 | 2.823 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
