
Hoeflea sp. IMCC20628
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Phyllobacteriaceae; Hoeflea; unclassified Hoeflea
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
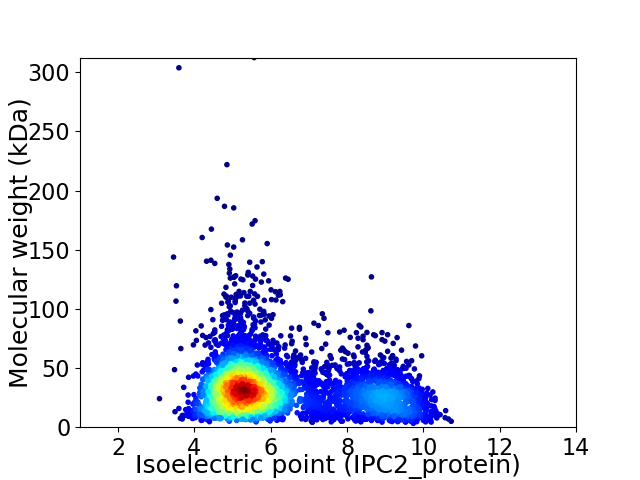
Virtual 2D-PAGE plot for 4739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7PMN7|A0A0F7PMN7_9RHIZ Universal stress protein UspA-like protein OS=Hoeflea sp. IMCC20628 OX=1620421 GN=IMCC20628_01705 PE=3 SV=1
MM1 pKa = 7.52SLLTDD6 pKa = 4.35SIDD9 pKa = 5.07AITFDD14 pKa = 4.87LGPDD18 pKa = 3.33EE19 pKa = 4.76TPIEE23 pKa = 4.57SINTSGASPYY33 pKa = 11.04VMNFQYY39 pKa = 11.09ASSQPADD46 pKa = 3.14LWNTYY51 pKa = 7.84TGWTSFSAAEE61 pKa = 3.9KK62 pKa = 8.75ATYY65 pKa = 9.82RR66 pKa = 11.84GLLDD70 pKa = 3.69YY71 pKa = 11.39VEE73 pKa = 4.32TLINVTFQEE82 pKa = 4.64VSGSSDD88 pKa = 3.17PDD90 pKa = 3.48MNVGKK95 pKa = 10.14VQIDD99 pKa = 3.5DD100 pKa = 3.83AAGKK104 pKa = 10.73GGFQYY109 pKa = 11.06YY110 pKa = 7.97YY111 pKa = 7.65TTSGQTATLTDD122 pKa = 3.68YY123 pKa = 11.47DD124 pKa = 3.95AFTVFEE130 pKa = 4.37NDD132 pKa = 2.64ISLEE136 pKa = 4.12SGSADD141 pKa = 4.68LILHH145 pKa = 6.82EE146 pKa = 5.39IGHH149 pKa = 6.96ALTLKK154 pKa = 10.5HH155 pKa = 6.46PFSGSPVLPTAYY167 pKa = 10.34DD168 pKa = 3.58NNKK171 pKa = 7.4YY172 pKa = 9.3TIMSYY177 pKa = 10.2TSNPDD182 pKa = 2.99TGLDD186 pKa = 3.55NEE188 pKa = 5.38GYY190 pKa = 10.47LAFDD194 pKa = 3.96IAALQQRR201 pKa = 11.84WGANMNTATGNDD213 pKa = 4.3VYY215 pKa = 10.34TGPRR219 pKa = 11.84IPTVDD224 pKa = 4.25VIWDD228 pKa = 3.41AGGTDD233 pKa = 3.37KK234 pKa = 11.33LSAFGYY240 pKa = 8.39STDD243 pKa = 3.46VLIDD247 pKa = 3.85LNEE250 pKa = 4.21GAFSRR255 pKa = 11.84FGTNDD260 pKa = 3.26DD261 pKa = 3.3VAIAYY266 pKa = 8.45GVVIEE271 pKa = 4.63NATGGNSHH279 pKa = 7.21DD280 pKa = 5.08LIIGNAQANTLAGGNGNDD298 pKa = 3.68NLSGGGGNDD307 pKa = 3.36YY308 pKa = 10.95FIGGNGIDD316 pKa = 4.05TYY318 pKa = 11.54DD319 pKa = 3.94GGSGSDD325 pKa = 3.13TARR328 pKa = 11.84FDD330 pKa = 4.75GNVGWRR336 pKa = 11.84VDD338 pKa = 4.08LGTGTAKK345 pKa = 10.54VGSQTEE351 pKa = 4.18MLTSIEE357 pKa = 4.16NVVGSNGNDD366 pKa = 3.66TITGTAYY373 pKa = 11.18SNDD376 pKa = 3.81LYY378 pKa = 11.3GGNGDD383 pKa = 4.68DD384 pKa = 3.98MLIGGGGIDD393 pKa = 4.52DD394 pKa = 4.55YY395 pKa = 11.75YY396 pKa = 11.29GGSGSDD402 pKa = 3.43TVDD405 pKa = 3.49LSGSNSKK412 pKa = 9.3WRR414 pKa = 11.84VDD416 pKa = 3.77LGNDD420 pKa = 3.37TAEE423 pKa = 4.23MTTATGNVYY432 pKa = 10.64NVDD435 pKa = 4.17FEE437 pKa = 5.11NIEE440 pKa = 4.35SVIGPRR446 pKa = 11.84NSLTLLGTSGNEE458 pKa = 3.74EE459 pKa = 3.6ARR461 pKa = 11.84GNSFADD467 pKa = 2.94IFYY470 pKa = 9.94TRR472 pKa = 11.84GGVDD476 pKa = 3.79EE477 pKa = 4.33FHH479 pKa = 7.0GSYY482 pKa = 11.14GSDD485 pKa = 2.68TVSFSGYY492 pKa = 10.99GEE494 pKa = 4.33GLTIRR499 pKa = 11.84GSAGYY504 pKa = 10.17GRR506 pKa = 11.84LGDD509 pKa = 3.88GTQVTTFTGIEE520 pKa = 4.2NIFATSEE527 pKa = 4.04DD528 pKa = 4.03DD529 pKa = 6.04DD530 pKa = 3.94ITGTAGNNTIVGRR543 pKa = 11.84SGADD547 pKa = 3.5RR548 pKa = 11.84INGGSGNDD556 pKa = 3.61IIQGGLGRR564 pKa = 11.84DD565 pKa = 3.61LLTGGRR571 pKa = 11.84GNDD574 pKa = 3.93TIDD577 pKa = 4.24GGSDD581 pKa = 3.23PNDD584 pKa = 3.14YY585 pKa = 11.53AFFSGNQSQYY595 pKa = 11.33NISTSAGGVTTVEE608 pKa = 4.27WIGPGNGDD616 pKa = 3.78GTDD619 pKa = 3.04TLTNIEE625 pKa = 4.67FLGFDD630 pKa = 3.38DD631 pKa = 4.33GFYY634 pKa = 11.03YY635 pKa = 10.73VV636 pKa = 4.22
MM1 pKa = 7.52SLLTDD6 pKa = 4.35SIDD9 pKa = 5.07AITFDD14 pKa = 4.87LGPDD18 pKa = 3.33EE19 pKa = 4.76TPIEE23 pKa = 4.57SINTSGASPYY33 pKa = 11.04VMNFQYY39 pKa = 11.09ASSQPADD46 pKa = 3.14LWNTYY51 pKa = 7.84TGWTSFSAAEE61 pKa = 3.9KK62 pKa = 8.75ATYY65 pKa = 9.82RR66 pKa = 11.84GLLDD70 pKa = 3.69YY71 pKa = 11.39VEE73 pKa = 4.32TLINVTFQEE82 pKa = 4.64VSGSSDD88 pKa = 3.17PDD90 pKa = 3.48MNVGKK95 pKa = 10.14VQIDD99 pKa = 3.5DD100 pKa = 3.83AAGKK104 pKa = 10.73GGFQYY109 pKa = 11.06YY110 pKa = 7.97YY111 pKa = 7.65TTSGQTATLTDD122 pKa = 3.68YY123 pKa = 11.47DD124 pKa = 3.95AFTVFEE130 pKa = 4.37NDD132 pKa = 2.64ISLEE136 pKa = 4.12SGSADD141 pKa = 4.68LILHH145 pKa = 6.82EE146 pKa = 5.39IGHH149 pKa = 6.96ALTLKK154 pKa = 10.5HH155 pKa = 6.46PFSGSPVLPTAYY167 pKa = 10.34DD168 pKa = 3.58NNKK171 pKa = 7.4YY172 pKa = 9.3TIMSYY177 pKa = 10.2TSNPDD182 pKa = 2.99TGLDD186 pKa = 3.55NEE188 pKa = 5.38GYY190 pKa = 10.47LAFDD194 pKa = 3.96IAALQQRR201 pKa = 11.84WGANMNTATGNDD213 pKa = 4.3VYY215 pKa = 10.34TGPRR219 pKa = 11.84IPTVDD224 pKa = 4.25VIWDD228 pKa = 3.41AGGTDD233 pKa = 3.37KK234 pKa = 11.33LSAFGYY240 pKa = 8.39STDD243 pKa = 3.46VLIDD247 pKa = 3.85LNEE250 pKa = 4.21GAFSRR255 pKa = 11.84FGTNDD260 pKa = 3.26DD261 pKa = 3.3VAIAYY266 pKa = 8.45GVVIEE271 pKa = 4.63NATGGNSHH279 pKa = 7.21DD280 pKa = 5.08LIIGNAQANTLAGGNGNDD298 pKa = 3.68NLSGGGGNDD307 pKa = 3.36YY308 pKa = 10.95FIGGNGIDD316 pKa = 4.05TYY318 pKa = 11.54DD319 pKa = 3.94GGSGSDD325 pKa = 3.13TARR328 pKa = 11.84FDD330 pKa = 4.75GNVGWRR336 pKa = 11.84VDD338 pKa = 4.08LGTGTAKK345 pKa = 10.54VGSQTEE351 pKa = 4.18MLTSIEE357 pKa = 4.16NVVGSNGNDD366 pKa = 3.66TITGTAYY373 pKa = 11.18SNDD376 pKa = 3.81LYY378 pKa = 11.3GGNGDD383 pKa = 4.68DD384 pKa = 3.98MLIGGGGIDD393 pKa = 4.52DD394 pKa = 4.55YY395 pKa = 11.75YY396 pKa = 11.29GGSGSDD402 pKa = 3.43TVDD405 pKa = 3.49LSGSNSKK412 pKa = 9.3WRR414 pKa = 11.84VDD416 pKa = 3.77LGNDD420 pKa = 3.37TAEE423 pKa = 4.23MTTATGNVYY432 pKa = 10.64NVDD435 pKa = 4.17FEE437 pKa = 5.11NIEE440 pKa = 4.35SVIGPRR446 pKa = 11.84NSLTLLGTSGNEE458 pKa = 3.74EE459 pKa = 3.6ARR461 pKa = 11.84GNSFADD467 pKa = 2.94IFYY470 pKa = 9.94TRR472 pKa = 11.84GGVDD476 pKa = 3.79EE477 pKa = 4.33FHH479 pKa = 7.0GSYY482 pKa = 11.14GSDD485 pKa = 2.68TVSFSGYY492 pKa = 10.99GEE494 pKa = 4.33GLTIRR499 pKa = 11.84GSAGYY504 pKa = 10.17GRR506 pKa = 11.84LGDD509 pKa = 3.88GTQVTTFTGIEE520 pKa = 4.2NIFATSEE527 pKa = 4.04DD528 pKa = 4.03DD529 pKa = 6.04DD530 pKa = 3.94ITGTAGNNTIVGRR543 pKa = 11.84SGADD547 pKa = 3.5RR548 pKa = 11.84INGGSGNDD556 pKa = 3.61IIQGGLGRR564 pKa = 11.84DD565 pKa = 3.61LLTGGRR571 pKa = 11.84GNDD574 pKa = 3.93TIDD577 pKa = 4.24GGSDD581 pKa = 3.23PNDD584 pKa = 3.14YY585 pKa = 11.53AFFSGNQSQYY595 pKa = 11.33NISTSAGGVTTVEE608 pKa = 4.27WIGPGNGDD616 pKa = 3.78GTDD619 pKa = 3.04TLTNIEE625 pKa = 4.67FLGFDD630 pKa = 3.38DD631 pKa = 4.33GFYY634 pKa = 11.03YY635 pKa = 10.73VV636 pKa = 4.22
Molecular weight: 66.48 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7PRS7|A0A0F7PRS7_9RHIZ Transcriptional regulator TetR family OS=Hoeflea sp. IMCC20628 OX=1620421 GN=IMCC20628_03186 PE=4 SV=1
MM1 pKa = 7.47TSGNRR6 pKa = 11.84LKK8 pKa = 10.85KK9 pKa = 10.18LRR11 pKa = 11.84KK12 pKa = 8.91QAAQRR17 pKa = 11.84QGGYY21 pKa = 10.08CFYY24 pKa = 10.61CRR26 pKa = 11.84YY27 pKa = 9.69PMWDD31 pKa = 3.16SRR33 pKa = 11.84PEE35 pKa = 3.99EE36 pKa = 4.09FIGRR40 pKa = 11.84YY41 pKa = 9.27GISSGLAQRR50 pKa = 11.84FQCTAEE56 pKa = 4.16HH57 pKa = 5.52VQARR61 pKa = 11.84CDD63 pKa = 3.5GGKK66 pKa = 10.04DD67 pKa = 3.28VAPNIVAACRR77 pKa = 11.84FCNSTRR83 pKa = 11.84HH84 pKa = 5.42KK85 pKa = 10.52AKK87 pKa = 10.27HH88 pKa = 5.58AADD91 pKa = 3.43AQTYY95 pKa = 8.65VSFVRR100 pKa = 11.84SRR102 pKa = 11.84LAKK105 pKa = 10.28GKK107 pKa = 7.66WHH109 pKa = 6.91PPQLRR114 pKa = 11.84ASFSEE119 pKa = 4.13RR120 pKa = 11.84KK121 pKa = 9.07NASDD125 pKa = 3.12RR126 pKa = 11.84SSRR129 pKa = 11.84ARR131 pKa = 3.29
MM1 pKa = 7.47TSGNRR6 pKa = 11.84LKK8 pKa = 10.85KK9 pKa = 10.18LRR11 pKa = 11.84KK12 pKa = 8.91QAAQRR17 pKa = 11.84QGGYY21 pKa = 10.08CFYY24 pKa = 10.61CRR26 pKa = 11.84YY27 pKa = 9.69PMWDD31 pKa = 3.16SRR33 pKa = 11.84PEE35 pKa = 3.99EE36 pKa = 4.09FIGRR40 pKa = 11.84YY41 pKa = 9.27GISSGLAQRR50 pKa = 11.84FQCTAEE56 pKa = 4.16HH57 pKa = 5.52VQARR61 pKa = 11.84CDD63 pKa = 3.5GGKK66 pKa = 10.04DD67 pKa = 3.28VAPNIVAACRR77 pKa = 11.84FCNSTRR83 pKa = 11.84HH84 pKa = 5.42KK85 pKa = 10.52AKK87 pKa = 10.27HH88 pKa = 5.58AADD91 pKa = 3.43AQTYY95 pKa = 8.65VSFVRR100 pKa = 11.84SRR102 pKa = 11.84LAKK105 pKa = 10.28GKK107 pKa = 7.66WHH109 pKa = 6.91PPQLRR114 pKa = 11.84ASFSEE119 pKa = 4.13RR120 pKa = 11.84KK121 pKa = 9.07NASDD125 pKa = 3.12RR126 pKa = 11.84SSRR129 pKa = 11.84ARR131 pKa = 3.29
Molecular weight: 14.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1486412 |
26 |
2994 |
313.7 |
34.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.712 ± 0.041 | 0.876 ± 0.011 |
5.948 ± 0.03 | 5.672 ± 0.036 |
3.934 ± 0.023 | 8.462 ± 0.039 |
2.048 ± 0.02 | 5.757 ± 0.03 |
3.625 ± 0.028 | 9.795 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.016 | 2.86 ± 0.02 |
4.851 ± 0.024 | 3.048 ± 0.018 |
6.277 ± 0.035 | 5.933 ± 0.025 |
5.511 ± 0.024 | 7.305 ± 0.03 |
1.315 ± 0.015 | 2.27 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
