
Pepper golden mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.46
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
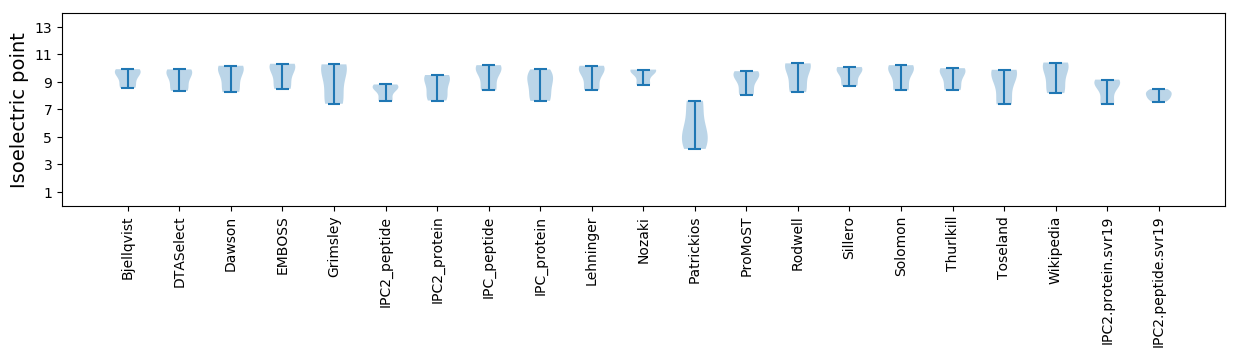
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q8QQA4|Q8QQA4_9GEMI Nuclear shuttle protein OS=Pepper golden mosaic virus OX=223301 GN=BR1 PE=3 SV=1
MM1 pKa = 7.34EE2 pKa = 6.2AGLASAPSAFNYY14 pKa = 10.39LEE16 pKa = 4.01SHH18 pKa = 6.52RR19 pKa = 11.84DD20 pKa = 3.56EE21 pKa = 4.67YY22 pKa = 11.24QLSHH26 pKa = 7.5DD27 pKa = 3.72LTEE30 pKa = 4.56IVLQFPSTASQLTARR45 pKa = 11.84LSRR48 pKa = 11.84SCMKK52 pKa = 9.84IDD54 pKa = 3.26HH55 pKa = 6.45CVIEE59 pKa = 4.31YY60 pKa = 9.79RR61 pKa = 11.84QQVPINASGSVIVEE75 pKa = 3.61IHH77 pKa = 6.0DD78 pKa = 3.94QRR80 pKa = 11.84MTDD83 pKa = 3.53NEE85 pKa = 4.36SLQASWTFPIRR96 pKa = 11.84CNIDD100 pKa = 2.83LHH102 pKa = 6.2YY103 pKa = 10.34FSSSFFSLKK112 pKa = 10.82DD113 pKa = 3.76PIPWRR118 pKa = 11.84LYY120 pKa = 10.45YY121 pKa = 10.11RR122 pKa = 11.84VSDD125 pKa = 3.96TNVHH129 pKa = 5.21QRR131 pKa = 11.84THH133 pKa = 5.37FAKK136 pKa = 10.85FKK138 pKa = 10.96GKK140 pKa = 10.52LKK142 pKa = 10.82LSTAKK147 pKa = 10.37HH148 pKa = 5.69SVDD151 pKa = 2.95IPFRR155 pKa = 11.84APTVKK160 pKa = 10.11ILSKK164 pKa = 10.54QFSHH168 pKa = 6.93NDD170 pKa = 2.75VDD172 pKa = 5.21FSHH175 pKa = 7.27VDD177 pKa = 3.14YY178 pKa = 10.91GQWEE182 pKa = 4.4RR183 pKa = 11.84KK184 pKa = 6.96VLRR187 pKa = 11.84SNSMSRR193 pKa = 11.84VGLTGPIEE201 pKa = 4.01LRR203 pKa = 11.84PGEE206 pKa = 4.29SWASRR211 pKa = 11.84STVGTSSPSRR221 pKa = 11.84DD222 pKa = 3.4SKK224 pKa = 8.38MHH226 pKa = 7.21PYY228 pKa = 10.29RR229 pKa = 11.84EE230 pKa = 4.07LHH232 pKa = 5.59QLGPSVLDD240 pKa = 4.4PGDD243 pKa = 3.79SASQVGLQRR252 pKa = 11.84AHH254 pKa = 6.7SNITMSMAQLNEE266 pKa = 4.01LVRR269 pKa = 11.84TTVHH273 pKa = 5.94EE274 pKa = 4.82CISNNCNPAQPKK286 pKa = 10.02SLQQ289 pKa = 3.27
MM1 pKa = 7.34EE2 pKa = 6.2AGLASAPSAFNYY14 pKa = 10.39LEE16 pKa = 4.01SHH18 pKa = 6.52RR19 pKa = 11.84DD20 pKa = 3.56EE21 pKa = 4.67YY22 pKa = 11.24QLSHH26 pKa = 7.5DD27 pKa = 3.72LTEE30 pKa = 4.56IVLQFPSTASQLTARR45 pKa = 11.84LSRR48 pKa = 11.84SCMKK52 pKa = 9.84IDD54 pKa = 3.26HH55 pKa = 6.45CVIEE59 pKa = 4.31YY60 pKa = 9.79RR61 pKa = 11.84QQVPINASGSVIVEE75 pKa = 3.61IHH77 pKa = 6.0DD78 pKa = 3.94QRR80 pKa = 11.84MTDD83 pKa = 3.53NEE85 pKa = 4.36SLQASWTFPIRR96 pKa = 11.84CNIDD100 pKa = 2.83LHH102 pKa = 6.2YY103 pKa = 10.34FSSSFFSLKK112 pKa = 10.82DD113 pKa = 3.76PIPWRR118 pKa = 11.84LYY120 pKa = 10.45YY121 pKa = 10.11RR122 pKa = 11.84VSDD125 pKa = 3.96TNVHH129 pKa = 5.21QRR131 pKa = 11.84THH133 pKa = 5.37FAKK136 pKa = 10.85FKK138 pKa = 10.96GKK140 pKa = 10.52LKK142 pKa = 10.82LSTAKK147 pKa = 10.37HH148 pKa = 5.69SVDD151 pKa = 2.95IPFRR155 pKa = 11.84APTVKK160 pKa = 10.11ILSKK164 pKa = 10.54QFSHH168 pKa = 6.93NDD170 pKa = 2.75VDD172 pKa = 5.21FSHH175 pKa = 7.27VDD177 pKa = 3.14YY178 pKa = 10.91GQWEE182 pKa = 4.4RR183 pKa = 11.84KK184 pKa = 6.96VLRR187 pKa = 11.84SNSMSRR193 pKa = 11.84VGLTGPIEE201 pKa = 4.01LRR203 pKa = 11.84PGEE206 pKa = 4.29SWASRR211 pKa = 11.84STVGTSSPSRR221 pKa = 11.84DD222 pKa = 3.4SKK224 pKa = 8.38MHH226 pKa = 7.21PYY228 pKa = 10.29RR229 pKa = 11.84EE230 pKa = 4.07LHH232 pKa = 5.59QLGPSVLDD240 pKa = 4.4PGDD243 pKa = 3.79SASQVGLQRR252 pKa = 11.84AHH254 pKa = 6.7SNITMSMAQLNEE266 pKa = 4.01LVRR269 pKa = 11.84TTVHH273 pKa = 5.94EE274 pKa = 4.82CISNNCNPAQPKK286 pKa = 10.02SLQQ289 pKa = 3.27
Molecular weight: 32.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q88884|Q88884_9GEMI Transcriptional activator protein OS=Pepper golden mosaic virus OX=223301 PE=3 SV=1
MM1 pKa = 8.25DD2 pKa = 4.26SRR4 pKa = 11.84TGEE7 pKa = 4.47SITVHH12 pKa = 4.76QAEE15 pKa = 4.03NSVFIWEE22 pKa = 4.39VPNPLFFRR30 pKa = 11.84IQHH33 pKa = 5.53VEE35 pKa = 3.99DD36 pKa = 3.8PLFTRR41 pKa = 11.84TRR43 pKa = 11.84IYY45 pKa = 10.43HH46 pKa = 4.83IVVRR50 pKa = 11.84FNHH53 pKa = 5.51NLRR56 pKa = 11.84RR57 pKa = 11.84ALALHH62 pKa = 6.42KK63 pKa = 10.65AFLHH67 pKa = 5.06FQVWTTSMTASGTTYY82 pKa = 10.72LSRR85 pKa = 11.84FKK87 pKa = 10.77FRR89 pKa = 11.84VMLYY93 pKa = 10.0LHH95 pKa = 6.33TLGAIGINNVIRR107 pKa = 11.84AVRR110 pKa = 11.84FATDD114 pKa = 3.02KK115 pKa = 11.32SYY117 pKa = 11.89VNYY120 pKa = 10.19VLEE123 pKa = 4.0NHH125 pKa = 6.63EE126 pKa = 4.81IKK128 pKa = 10.75CKK130 pKa = 10.61LYY132 pKa = 10.81
MM1 pKa = 8.25DD2 pKa = 4.26SRR4 pKa = 11.84TGEE7 pKa = 4.47SITVHH12 pKa = 4.76QAEE15 pKa = 4.03NSVFIWEE22 pKa = 4.39VPNPLFFRR30 pKa = 11.84IQHH33 pKa = 5.53VEE35 pKa = 3.99DD36 pKa = 3.8PLFTRR41 pKa = 11.84TRR43 pKa = 11.84IYY45 pKa = 10.43HH46 pKa = 4.83IVVRR50 pKa = 11.84FNHH53 pKa = 5.51NLRR56 pKa = 11.84RR57 pKa = 11.84ALALHH62 pKa = 6.42KK63 pKa = 10.65AFLHH67 pKa = 5.06FQVWTTSMTASGTTYY82 pKa = 10.72LSRR85 pKa = 11.84FKK87 pKa = 10.77FRR89 pKa = 11.84VMLYY93 pKa = 10.0LHH95 pKa = 6.33TLGAIGINNVIRR107 pKa = 11.84AVRR110 pKa = 11.84FATDD114 pKa = 3.02KK115 pKa = 11.32SYY117 pKa = 11.89VNYY120 pKa = 10.19VLEE123 pKa = 4.0NHH125 pKa = 6.63EE126 pKa = 4.81IKK128 pKa = 10.75CKK130 pKa = 10.61LYY132 pKa = 10.81
Molecular weight: 15.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1405 |
129 |
348 |
234.2 |
26.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.836 ± 0.67 | 2.064 ± 0.211 |
5.053 ± 0.371 | 3.986 ± 0.349 |
4.555 ± 0.5 | 5.053 ± 0.412 |
4.128 ± 0.371 | 5.694 ± 0.553 |
6.05 ± 0.739 | 6.477 ± 0.626 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.407 | 5.623 ± 0.406 |
5.267 ± 0.823 | 3.843 ± 0.462 |
6.975 ± 0.894 | 8.968 ± 1.296 |
4.982 ± 0.543 | 7.189 ± 1.036 |
1.495 ± 0.225 | 4.27 ± 0.586 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
