
Capybara microvirus Cap1_SP_82
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
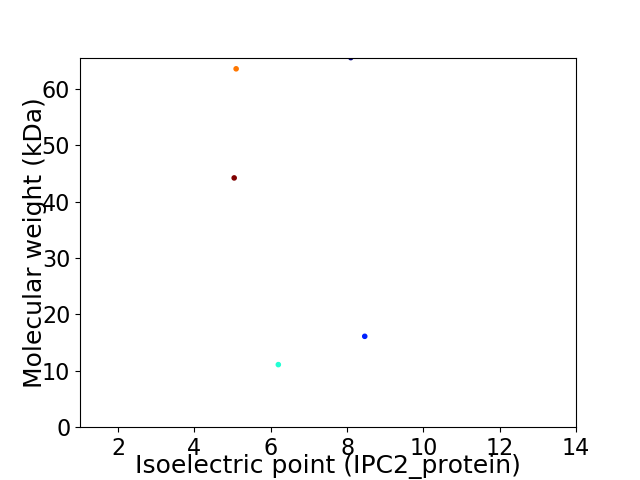
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVN2|A0A4V1FVN2_9VIRU Replication initiator protein OS=Capybara microvirus Cap1_SP_82 OX=2584793 PE=4 SV=1
MM1 pKa = 7.63SIFDD5 pKa = 3.3IHH7 pKa = 6.77KK8 pKa = 10.3LKK10 pKa = 10.98NSPTWSEE17 pKa = 3.75FDD19 pKa = 2.81ISQNKK24 pKa = 8.95KK25 pKa = 8.66FSSKK29 pKa = 9.82IGQILPVAYY38 pKa = 8.56RR39 pKa = 11.84TVLPGDD45 pKa = 3.84KK46 pKa = 10.56VNLPISHH53 pKa = 7.0FTQTLPLNKK62 pKa = 9.91NSFARR67 pKa = 11.84IRR69 pKa = 11.84EE70 pKa = 4.12HH71 pKa = 7.77VDD73 pKa = 2.86VFFVPFRR80 pKa = 11.84LLYY83 pKa = 10.87SKK85 pKa = 11.43ANDD88 pKa = 2.97ILVNNTDD95 pKa = 3.47SQVALANSATNNIIPTEE112 pKa = 4.11LPFTTISILNSAYY125 pKa = 9.78KK126 pKa = 10.29FYY128 pKa = 10.58KK129 pKa = 10.5AEE131 pKa = 4.09EE132 pKa = 4.24ASPSYY137 pKa = 9.68TFKK140 pKa = 11.01GHH142 pKa = 7.46DD143 pKa = 2.93IAGFSRR149 pKa = 11.84AEE151 pKa = 3.76NMKK154 pKa = 9.93YY155 pKa = 10.2LCEE158 pKa = 3.84LLGYY162 pKa = 10.95GDD164 pKa = 3.57ITRR167 pKa = 11.84GYY169 pKa = 9.73ADD171 pKa = 3.82SEE173 pKa = 4.54GSVDD177 pKa = 3.97NKK179 pKa = 10.96AHH181 pKa = 6.78IPLSMMPICAYY192 pKa = 10.1QKK194 pKa = 10.01IYY196 pKa = 11.32ADD198 pKa = 3.89FFRR201 pKa = 11.84YY202 pKa = 7.74QQWEE206 pKa = 4.21DD207 pKa = 3.62NNPYY211 pKa = 9.85TYY213 pKa = 11.33NFDD216 pKa = 3.57YY217 pKa = 10.87KK218 pKa = 10.38IASNGMQVSLQEE230 pKa = 3.51ITLPGFGANIYY241 pKa = 10.96GNGIFDD247 pKa = 5.2LRR249 pKa = 11.84YY250 pKa = 9.33KK251 pKa = 10.67NYY253 pKa = 10.71NKK255 pKa = 10.45DD256 pKa = 3.51YY257 pKa = 10.95YY258 pKa = 10.96FGLLPSQVNDD268 pKa = 2.81IYY270 pKa = 11.26LPKK273 pKa = 10.24FDD275 pKa = 4.31MTLDD279 pKa = 3.63TEE281 pKa = 4.32KK282 pKa = 10.45ATEE285 pKa = 3.98EE286 pKa = 4.09GHH288 pKa = 5.23FQRR291 pKa = 11.84FNDD294 pKa = 3.09ITTTLSVLQLRR305 pKa = 11.84ASYY308 pKa = 10.98ALQKK312 pKa = 10.01WSEE315 pKa = 3.96IQLFSDD321 pKa = 4.15KK322 pKa = 10.89NYY324 pKa = 9.84KK325 pKa = 10.44DD326 pKa = 3.66KK327 pKa = 10.93IKK329 pKa = 10.44HH330 pKa = 5.75HH331 pKa = 6.78FNVDD335 pKa = 3.34VPDD338 pKa = 3.68YY339 pKa = 9.47TANQAQYY346 pKa = 10.45LGGYY350 pKa = 8.77SSVMQINDD358 pKa = 3.17IVNTNLAQANAEE370 pKa = 3.98RR371 pKa = 11.84KK372 pKa = 10.18AIGNSANSGHH382 pKa = 6.89INFDD386 pKa = 3.9CKK388 pKa = 10.32EE389 pKa = 4.1HH390 pKa = 6.63GIIMAVYY397 pKa = 8.88YY398 pKa = 10.12ALPLMDD404 pKa = 4.71YY405 pKa = 8.07GTLYY409 pKa = 10.86GYY411 pKa = 10.57DD412 pKa = 3.6PEE414 pKa = 4.63VTHH417 pKa = 5.73VTADD421 pKa = 3.84DD422 pKa = 4.03FPKK425 pKa = 10.79PEE427 pKa = 4.29FDD429 pKa = 4.18NLGLSPVYY437 pKa = 10.2LHH439 pKa = 6.6NLIDD443 pKa = 4.46VPDD446 pKa = 4.36NLDD449 pKa = 3.38KK450 pKa = 11.08PIGYY454 pKa = 9.88APRR457 pKa = 11.84YY458 pKa = 8.49WDD460 pKa = 4.31FKK462 pKa = 10.29SCVDD466 pKa = 4.12KK467 pKa = 11.7YY468 pKa = 11.16CGAFNLEE475 pKa = 3.98QFRR478 pKa = 11.84GFTIEE483 pKa = 4.51HH484 pKa = 6.92PWNLLYY490 pKa = 10.68DD491 pKa = 4.01IALSEE496 pKa = 4.38HH497 pKa = 7.09PEE499 pKa = 4.32DD500 pKa = 5.39LPDD503 pKa = 4.15IYY505 pKa = 10.98WMALFKK511 pKa = 10.02CTPNDD516 pKa = 3.2AKK518 pKa = 10.91FIFSALPDD526 pKa = 3.75STRR529 pKa = 11.84LTDD532 pKa = 3.52TFLTSLDD539 pKa = 3.25FDD541 pKa = 4.57FKK543 pKa = 11.47VNRR546 pKa = 11.84CLSYY550 pKa = 11.35NGMPYY555 pKa = 10.91
MM1 pKa = 7.63SIFDD5 pKa = 3.3IHH7 pKa = 6.77KK8 pKa = 10.3LKK10 pKa = 10.98NSPTWSEE17 pKa = 3.75FDD19 pKa = 2.81ISQNKK24 pKa = 8.95KK25 pKa = 8.66FSSKK29 pKa = 9.82IGQILPVAYY38 pKa = 8.56RR39 pKa = 11.84TVLPGDD45 pKa = 3.84KK46 pKa = 10.56VNLPISHH53 pKa = 7.0FTQTLPLNKK62 pKa = 9.91NSFARR67 pKa = 11.84IRR69 pKa = 11.84EE70 pKa = 4.12HH71 pKa = 7.77VDD73 pKa = 2.86VFFVPFRR80 pKa = 11.84LLYY83 pKa = 10.87SKK85 pKa = 11.43ANDD88 pKa = 2.97ILVNNTDD95 pKa = 3.47SQVALANSATNNIIPTEE112 pKa = 4.11LPFTTISILNSAYY125 pKa = 9.78KK126 pKa = 10.29FYY128 pKa = 10.58KK129 pKa = 10.5AEE131 pKa = 4.09EE132 pKa = 4.24ASPSYY137 pKa = 9.68TFKK140 pKa = 11.01GHH142 pKa = 7.46DD143 pKa = 2.93IAGFSRR149 pKa = 11.84AEE151 pKa = 3.76NMKK154 pKa = 9.93YY155 pKa = 10.2LCEE158 pKa = 3.84LLGYY162 pKa = 10.95GDD164 pKa = 3.57ITRR167 pKa = 11.84GYY169 pKa = 9.73ADD171 pKa = 3.82SEE173 pKa = 4.54GSVDD177 pKa = 3.97NKK179 pKa = 10.96AHH181 pKa = 6.78IPLSMMPICAYY192 pKa = 10.1QKK194 pKa = 10.01IYY196 pKa = 11.32ADD198 pKa = 3.89FFRR201 pKa = 11.84YY202 pKa = 7.74QQWEE206 pKa = 4.21DD207 pKa = 3.62NNPYY211 pKa = 9.85TYY213 pKa = 11.33NFDD216 pKa = 3.57YY217 pKa = 10.87KK218 pKa = 10.38IASNGMQVSLQEE230 pKa = 3.51ITLPGFGANIYY241 pKa = 10.96GNGIFDD247 pKa = 5.2LRR249 pKa = 11.84YY250 pKa = 9.33KK251 pKa = 10.67NYY253 pKa = 10.71NKK255 pKa = 10.45DD256 pKa = 3.51YY257 pKa = 10.95YY258 pKa = 10.96FGLLPSQVNDD268 pKa = 2.81IYY270 pKa = 11.26LPKK273 pKa = 10.24FDD275 pKa = 4.31MTLDD279 pKa = 3.63TEE281 pKa = 4.32KK282 pKa = 10.45ATEE285 pKa = 3.98EE286 pKa = 4.09GHH288 pKa = 5.23FQRR291 pKa = 11.84FNDD294 pKa = 3.09ITTTLSVLQLRR305 pKa = 11.84ASYY308 pKa = 10.98ALQKK312 pKa = 10.01WSEE315 pKa = 3.96IQLFSDD321 pKa = 4.15KK322 pKa = 10.89NYY324 pKa = 9.84KK325 pKa = 10.44DD326 pKa = 3.66KK327 pKa = 10.93IKK329 pKa = 10.44HH330 pKa = 5.75HH331 pKa = 6.78FNVDD335 pKa = 3.34VPDD338 pKa = 3.68YY339 pKa = 9.47TANQAQYY346 pKa = 10.45LGGYY350 pKa = 8.77SSVMQINDD358 pKa = 3.17IVNTNLAQANAEE370 pKa = 3.98RR371 pKa = 11.84KK372 pKa = 10.18AIGNSANSGHH382 pKa = 6.89INFDD386 pKa = 3.9CKK388 pKa = 10.32EE389 pKa = 4.1HH390 pKa = 6.63GIIMAVYY397 pKa = 8.88YY398 pKa = 10.12ALPLMDD404 pKa = 4.71YY405 pKa = 8.07GTLYY409 pKa = 10.86GYY411 pKa = 10.57DD412 pKa = 3.6PEE414 pKa = 4.63VTHH417 pKa = 5.73VTADD421 pKa = 3.84DD422 pKa = 4.03FPKK425 pKa = 10.79PEE427 pKa = 4.29FDD429 pKa = 4.18NLGLSPVYY437 pKa = 10.2LHH439 pKa = 6.6NLIDD443 pKa = 4.46VPDD446 pKa = 4.36NLDD449 pKa = 3.38KK450 pKa = 11.08PIGYY454 pKa = 9.88APRR457 pKa = 11.84YY458 pKa = 8.49WDD460 pKa = 4.31FKK462 pKa = 10.29SCVDD466 pKa = 4.12KK467 pKa = 11.7YY468 pKa = 11.16CGAFNLEE475 pKa = 3.98QFRR478 pKa = 11.84GFTIEE483 pKa = 4.51HH484 pKa = 6.92PWNLLYY490 pKa = 10.68DD491 pKa = 4.01IALSEE496 pKa = 4.38HH497 pKa = 7.09PEE499 pKa = 4.32DD500 pKa = 5.39LPDD503 pKa = 4.15IYY505 pKa = 10.98WMALFKK511 pKa = 10.02CTPNDD516 pKa = 3.2AKK518 pKa = 10.91FIFSALPDD526 pKa = 3.75STRR529 pKa = 11.84LTDD532 pKa = 3.52TFLTSLDD539 pKa = 3.25FDD541 pKa = 4.57FKK543 pKa = 11.47VNRR546 pKa = 11.84CLSYY550 pKa = 11.35NGMPYY555 pKa = 10.91
Molecular weight: 63.59 kDa
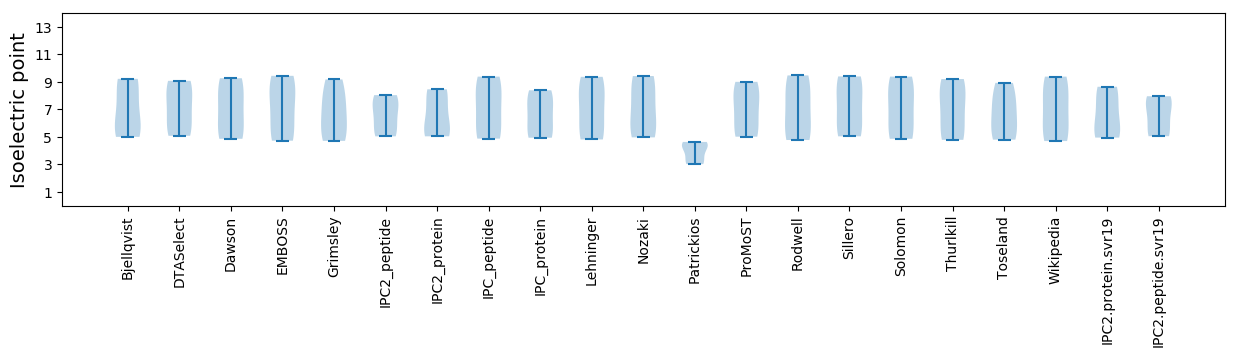
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W404|A0A4P8W404_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_82 OX=2584793 PE=3 SV=1
MM1 pKa = 7.78CIILNPKK8 pKa = 8.0TQLSEE13 pKa = 4.22HH14 pKa = 6.42FSLTEE19 pKa = 3.87MVSKK23 pKa = 10.85NDD25 pKa = 3.52PCHH28 pKa = 7.11DD29 pKa = 3.85VLLKK33 pKa = 10.44KK34 pKa = 10.78LPVHH38 pKa = 6.67VYY40 pKa = 9.73EE41 pKa = 4.49NLKK44 pKa = 10.1KK45 pKa = 10.06VCSILEE51 pKa = 4.18EE52 pKa = 3.9FRR54 pKa = 11.84KK55 pKa = 10.2ILDD58 pKa = 3.68CPISVNSALRR68 pKa = 11.84STSHH72 pKa = 6.65NIAVGGVTNSYY83 pKa = 9.25HH84 pKa = 5.65CHH86 pKa = 5.41GLAVDD91 pKa = 4.47FRR93 pKa = 11.84TNKK96 pKa = 8.68TALSNFQLLSSAYY109 pKa = 9.34NLKK112 pKa = 10.21PYY114 pKa = 10.58KK115 pKa = 10.29SGNYY119 pKa = 9.28LVAKK123 pKa = 9.91NDD125 pKa = 3.39KK126 pKa = 10.35FKK128 pKa = 11.37LLLYY132 pKa = 7.68PTFIHH137 pKa = 6.1MQLQKK142 pKa = 11.18
MM1 pKa = 7.78CIILNPKK8 pKa = 8.0TQLSEE13 pKa = 4.22HH14 pKa = 6.42FSLTEE19 pKa = 3.87MVSKK23 pKa = 10.85NDD25 pKa = 3.52PCHH28 pKa = 7.11DD29 pKa = 3.85VLLKK33 pKa = 10.44KK34 pKa = 10.78LPVHH38 pKa = 6.67VYY40 pKa = 9.73EE41 pKa = 4.49NLKK44 pKa = 10.1KK45 pKa = 10.06VCSILEE51 pKa = 4.18EE52 pKa = 3.9FRR54 pKa = 11.84KK55 pKa = 10.2ILDD58 pKa = 3.68CPISVNSALRR68 pKa = 11.84STSHH72 pKa = 6.65NIAVGGVTNSYY83 pKa = 9.25HH84 pKa = 5.65CHH86 pKa = 5.41GLAVDD91 pKa = 4.47FRR93 pKa = 11.84TNKK96 pKa = 8.68TALSNFQLLSSAYY109 pKa = 9.34NLKK112 pKa = 10.21PYY114 pKa = 10.58KK115 pKa = 10.29SGNYY119 pKa = 9.28LVAKK123 pKa = 9.91NDD125 pKa = 3.39KK126 pKa = 10.35FKK128 pKa = 11.37LLLYY132 pKa = 7.68PTFIHH137 pKa = 6.1MQLQKK142 pKa = 11.18
Molecular weight: 16.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1734 |
98 |
555 |
346.8 |
40.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.306 ± 0.595 | 1.557 ± 0.447 |
5.536 ± 0.855 | 5.421 ± 0.811 |
5.709 ± 1.133 | 3.922 ± 0.457 |
2.307 ± 0.636 | 6.171 ± 0.342 |
6.92 ± 0.508 | 10.092 ± 0.763 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.73 ± 0.228 | 8.939 ± 1.005 |
4.325 ± 0.732 | 5.306 ± 1.858 |
3.633 ± 0.482 | 6.69 ± 0.467 |
5.421 ± 0.154 | 4.268 ± 0.433 |
0.577 ± 0.213 | 6.171 ± 1.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
