
Salmonella sp. NCTC 7297
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Salmonella; unclassified Salmonella
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
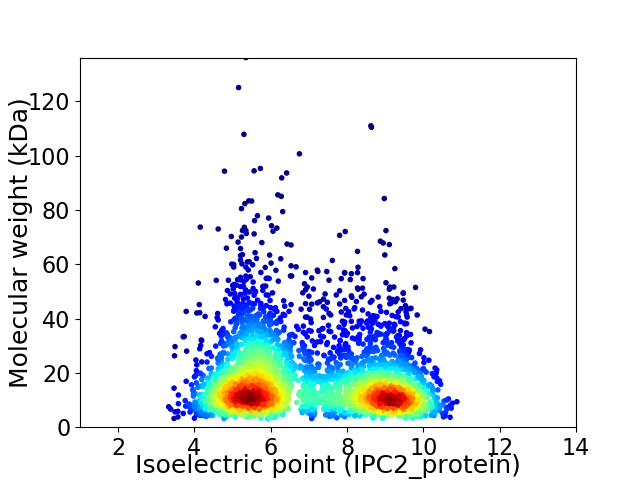
Virtual 2D-PAGE plot for 3497 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A509D1S3|A0A509D1S3_9ENTR T3SS secreted effector EspN-like protein OS=Salmonella sp. NCTC 7297 OX=2583578 GN=NCTC7297_02951 PE=4 SV=1
MM1 pKa = 7.67LEE3 pKa = 4.0QMEE6 pKa = 4.47LTDD9 pKa = 4.16YY10 pKa = 11.88ANMALSIPSANTNIWNLEE28 pKa = 3.6QDD30 pKa = 3.86TVGTRR35 pKa = 11.84LTNARR40 pKa = 11.84HH41 pKa = 5.4GLADD45 pKa = 4.02NGGAWVSYY53 pKa = 10.43FGGNFNGDD61 pKa = 3.55NGTINYY67 pKa = 8.82DD68 pKa = 3.13QDD70 pKa = 3.61VNGIMVGVDD79 pKa = 2.99TKK81 pKa = 11.4VDD83 pKa = 3.43GNNAKK88 pKa = 9.88WIVGAAAGFAKK99 pKa = 10.49GDD101 pKa = 3.51LSDD104 pKa = 3.46RR105 pKa = 11.84TGQVDD110 pKa = 3.54QDD112 pKa = 3.84SQSAYY117 pKa = 9.05IYY119 pKa = 10.73SSAHH123 pKa = 4.91FANNIFVDD131 pKa = 4.18GNLSYY136 pKa = 10.95FHH138 pKa = 7.14FNNDD142 pKa = 2.59LSANMSDD149 pKa = 3.19GTYY152 pKa = 10.73VDD154 pKa = 5.14GNTSSDD160 pKa = 2.69AWGFGLKK167 pKa = 10.32LGYY170 pKa = 9.47DD171 pKa = 3.67WKK173 pKa = 11.2LGDD176 pKa = 4.4AGYY179 pKa = 7.83LTPYY183 pKa = 10.28GSVSGLFQSGDD194 pKa = 3.79DD195 pKa = 3.89YY196 pKa = 11.83QLSNDD201 pKa = 3.96MKK203 pKa = 11.23VDD205 pKa = 3.62GQSYY209 pKa = 10.64DD210 pKa = 3.02SMRR213 pKa = 11.84YY214 pKa = 8.94EE215 pKa = 4.34LGVDD219 pKa = 3.21AGYY222 pKa = 8.52TFTYY226 pKa = 10.71SDD228 pKa = 4.19DD229 pKa = 3.86QALTPYY235 pKa = 10.29FKK237 pKa = 10.64LAYY240 pKa = 10.52VYY242 pKa = 11.0DD243 pKa = 4.56DD244 pKa = 4.43SNNDD248 pKa = 3.06ADD250 pKa = 4.41VNGDD254 pKa = 3.84SIDD257 pKa = 3.7NGVEE261 pKa = 3.97GSAVRR266 pKa = 11.84VGLGTQFSFTKK277 pKa = 10.49NFSAYY282 pKa = 9.3TDD284 pKa = 3.45ANYY287 pKa = 10.81LGGGDD292 pKa = 4.6VDD294 pKa = 4.32QDD296 pKa = 2.67WSANVGVKK304 pKa = 7.43YY305 pKa = 8.84TWW307 pKa = 3.09
MM1 pKa = 7.67LEE3 pKa = 4.0QMEE6 pKa = 4.47LTDD9 pKa = 4.16YY10 pKa = 11.88ANMALSIPSANTNIWNLEE28 pKa = 3.6QDD30 pKa = 3.86TVGTRR35 pKa = 11.84LTNARR40 pKa = 11.84HH41 pKa = 5.4GLADD45 pKa = 4.02NGGAWVSYY53 pKa = 10.43FGGNFNGDD61 pKa = 3.55NGTINYY67 pKa = 8.82DD68 pKa = 3.13QDD70 pKa = 3.61VNGIMVGVDD79 pKa = 2.99TKK81 pKa = 11.4VDD83 pKa = 3.43GNNAKK88 pKa = 9.88WIVGAAAGFAKK99 pKa = 10.49GDD101 pKa = 3.51LSDD104 pKa = 3.46RR105 pKa = 11.84TGQVDD110 pKa = 3.54QDD112 pKa = 3.84SQSAYY117 pKa = 9.05IYY119 pKa = 10.73SSAHH123 pKa = 4.91FANNIFVDD131 pKa = 4.18GNLSYY136 pKa = 10.95FHH138 pKa = 7.14FNNDD142 pKa = 2.59LSANMSDD149 pKa = 3.19GTYY152 pKa = 10.73VDD154 pKa = 5.14GNTSSDD160 pKa = 2.69AWGFGLKK167 pKa = 10.32LGYY170 pKa = 9.47DD171 pKa = 3.67WKK173 pKa = 11.2LGDD176 pKa = 4.4AGYY179 pKa = 7.83LTPYY183 pKa = 10.28GSVSGLFQSGDD194 pKa = 3.79DD195 pKa = 3.89YY196 pKa = 11.83QLSNDD201 pKa = 3.96MKK203 pKa = 11.23VDD205 pKa = 3.62GQSYY209 pKa = 10.64DD210 pKa = 3.02SMRR213 pKa = 11.84YY214 pKa = 8.94EE215 pKa = 4.34LGVDD219 pKa = 3.21AGYY222 pKa = 8.52TFTYY226 pKa = 10.71SDD228 pKa = 4.19DD229 pKa = 3.86QALTPYY235 pKa = 10.29FKK237 pKa = 10.64LAYY240 pKa = 10.52VYY242 pKa = 11.0DD243 pKa = 4.56DD244 pKa = 4.43SNNDD248 pKa = 3.06ADD250 pKa = 4.41VNGDD254 pKa = 3.84SIDD257 pKa = 3.7NGVEE261 pKa = 3.97GSAVRR266 pKa = 11.84VGLGTQFSFTKK277 pKa = 10.49NFSAYY282 pKa = 9.3TDD284 pKa = 3.45ANYY287 pKa = 10.81LGGGDD292 pKa = 4.6VDD294 pKa = 4.32QDD296 pKa = 2.67WSANVGVKK304 pKa = 7.43YY305 pKa = 8.84TWW307 pKa = 3.09
Molecular weight: 33.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A509BFQ1|A0A509BFQ1_9ENTR Endolysin OS=Salmonella sp. NCTC 7297 OX=2583578 GN=NCTC7297_00326 PE=4 SV=1
MM1 pKa = 7.4NRR3 pKa = 11.84QSWLLNLSLLKK14 pKa = 8.97THH16 pKa = 6.9PAFRR20 pKa = 11.84AVFLARR26 pKa = 11.84FISIVSLGLLGVAVPVQIQMMTHH49 pKa = 5.51STWQVGLSVTLTGGAMFIGLMVGGVLADD77 pKa = 3.64RR78 pKa = 11.84YY79 pKa = 9.17EE80 pKa = 4.1RR81 pKa = 11.84KK82 pKa = 9.81KK83 pKa = 11.2VILLARR89 pKa = 11.84GTCGVGFIGLCVNALLPEE107 pKa = 4.42PSLLAIYY114 pKa = 10.65LLGLWDD120 pKa = 4.55GFFASLGRR128 pKa = 11.84DD129 pKa = 2.68RR130 pKa = 11.84FYY132 pKa = 10.59WRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84QRR138 pKa = 11.84WSEE141 pKa = 3.76EE142 pKa = 4.14KK143 pKa = 10.07ICCRR147 pKa = 11.84PARR150 pKa = 11.84LPCC153 pKa = 4.6
MM1 pKa = 7.4NRR3 pKa = 11.84QSWLLNLSLLKK14 pKa = 8.97THH16 pKa = 6.9PAFRR20 pKa = 11.84AVFLARR26 pKa = 11.84FISIVSLGLLGVAVPVQIQMMTHH49 pKa = 5.51STWQVGLSVTLTGGAMFIGLMVGGVLADD77 pKa = 3.64RR78 pKa = 11.84YY79 pKa = 9.17EE80 pKa = 4.1RR81 pKa = 11.84KK82 pKa = 9.81KK83 pKa = 11.2VILLARR89 pKa = 11.84GTCGVGFIGLCVNALLPEE107 pKa = 4.42PSLLAIYY114 pKa = 10.65LLGLWDD120 pKa = 4.55GFFASLGRR128 pKa = 11.84DD129 pKa = 2.68RR130 pKa = 11.84FYY132 pKa = 10.59WRR134 pKa = 11.84RR135 pKa = 11.84RR136 pKa = 11.84QRR138 pKa = 11.84WSEE141 pKa = 3.76EE142 pKa = 4.14KK143 pKa = 10.07ICCRR147 pKa = 11.84PARR150 pKa = 11.84LPCC153 pKa = 4.6
Molecular weight: 17.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
599048 |
29 |
1259 |
171.3 |
19.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.59 ± 0.051 | 1.298 ± 0.02 |
5.085 ± 0.037 | 5.282 ± 0.045 |
3.889 ± 0.036 | 7.312 ± 0.043 |
2.336 ± 0.028 | 5.932 ± 0.047 |
4.255 ± 0.038 | 10.415 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.978 ± 0.027 | 3.84 ± 0.038 |
4.533 ± 0.037 | 4.288 ± 0.039 |
6.219 ± 0.046 | 5.968 ± 0.04 |
5.532 ± 0.042 | 6.814 ± 0.044 |
1.573 ± 0.023 | 2.862 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
