
Rubellimicrobium mesophilum DSM 19309
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Rubellimicrobium; Rubellimicrobium mesophilum
Average proteome isoelectric point is 6.47
Get precalculated fractions of proteins
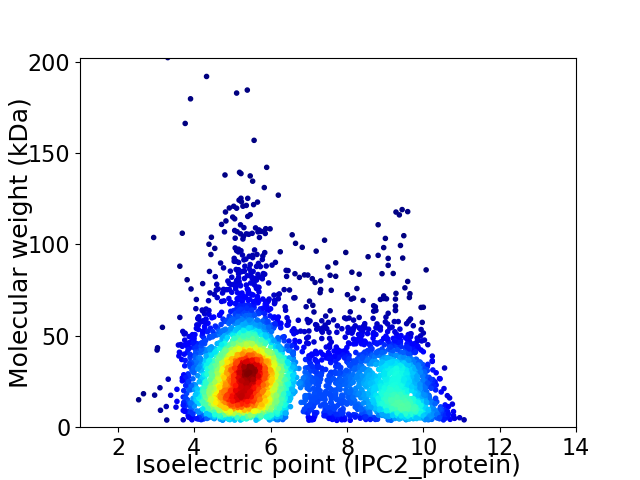
Virtual 2D-PAGE plot for 4946 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A017HIU9|A0A017HIU9_9RHOB GTA NlpC/P60 family peptidase OS=Rubellimicrobium mesophilum DSM 19309 OX=442562 GN=Rumeso_03964 PE=3 SV=1
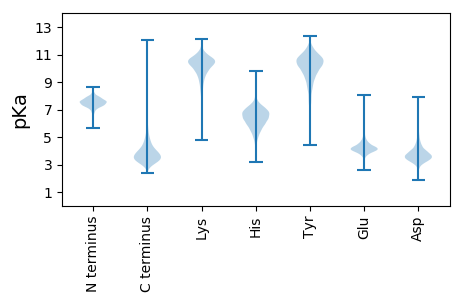
MM1 pKa = 7.3TRR3 pKa = 11.84LTTATALALIWASGALAQTEE23 pKa = 4.3LTMWYY28 pKa = 9.48HH29 pKa = 5.91GAGNQVEE36 pKa = 4.48SEE38 pKa = 4.66IINQIISDD46 pKa = 4.13FNASQSDD53 pKa = 3.24WHH55 pKa = 5.47VTIEE59 pKa = 4.34SFPQAAYY66 pKa = 10.8NDD68 pKa = 4.13AVVAAALAGDD78 pKa = 4.59LPDD81 pKa = 4.6ILDD84 pKa = 3.42VDD86 pKa = 4.67GPIMPNWAWAGYY98 pKa = 7.77LQPLPIDD105 pKa = 3.61PAKK108 pKa = 10.48IEE110 pKa = 4.25SFLPGPKK117 pKa = 9.13GTWNDD122 pKa = 3.15QLYY125 pKa = 10.91SVGLWDD131 pKa = 4.01AAVALVTRR139 pKa = 11.84QSTLDD144 pKa = 3.53EE145 pKa = 4.93LGLRR149 pKa = 11.84TPTLEE154 pKa = 4.19EE155 pKa = 4.0PWTLEE160 pKa = 3.76EE161 pKa = 5.23FMGAMEE167 pKa = 4.41AAKK170 pKa = 10.64ASGKK174 pKa = 10.05YY175 pKa = 10.04DD176 pKa = 3.5YY177 pKa = 11.18AIDD180 pKa = 5.27LGMSDD185 pKa = 3.01QGEE188 pKa = 4.35WYY190 pKa = 9.03PYY192 pKa = 11.22AFLPFLWSFGGDD204 pKa = 3.37LVDD207 pKa = 4.5VQADD211 pKa = 3.6PQTAEE216 pKa = 4.01GVLNGDD222 pKa = 3.69AGLKK226 pKa = 9.89FGEE229 pKa = 4.02WWQGMFANGYY239 pKa = 10.37APGPSQDD246 pKa = 4.13PADD249 pKa = 4.04HH250 pKa = 7.12ASGFQEE256 pKa = 3.99GKK258 pKa = 10.64YY259 pKa = 10.04AFSWNGNWLALPFLEE274 pKa = 6.02AYY276 pKa = 10.29DD277 pKa = 4.14DD278 pKa = 4.38VVFLPAPDD286 pKa = 4.4FGNGNYY292 pKa = 9.6IGGASWQFAVSGTTEE307 pKa = 3.99HH308 pKa = 7.35PEE310 pKa = 3.73GGAAFIEE317 pKa = 4.86FALQDD322 pKa = 3.24KK323 pKa = 10.5YY324 pKa = 11.16LAQFSDD330 pKa = 5.05GIGLIPPTTTAAAMTQNYY348 pKa = 9.13KK349 pKa = 10.57EE350 pKa = 4.35SGPMAVFYY358 pKa = 10.81DD359 pKa = 4.38LSADD363 pKa = 3.67QARR366 pKa = 11.84LRR368 pKa = 11.84PVTPGYY374 pKa = 9.65PVASKK379 pKa = 11.12VFLKK383 pKa = 11.21AMMDD387 pKa = 3.45IANGADD393 pKa = 3.34VADD396 pKa = 4.32TLDD399 pKa = 3.68SAVDD403 pKa = 4.27EE404 pKa = 4.58IDD406 pKa = 5.5ADD408 pKa = 3.53IQANNGYY415 pKa = 9.69AQAQQ419 pKa = 3.46
MM1 pKa = 7.3TRR3 pKa = 11.84LTTATALALIWASGALAQTEE23 pKa = 4.3LTMWYY28 pKa = 9.48HH29 pKa = 5.91GAGNQVEE36 pKa = 4.48SEE38 pKa = 4.66IINQIISDD46 pKa = 4.13FNASQSDD53 pKa = 3.24WHH55 pKa = 5.47VTIEE59 pKa = 4.34SFPQAAYY66 pKa = 10.8NDD68 pKa = 4.13AVVAAALAGDD78 pKa = 4.59LPDD81 pKa = 4.6ILDD84 pKa = 3.42VDD86 pKa = 4.67GPIMPNWAWAGYY98 pKa = 7.77LQPLPIDD105 pKa = 3.61PAKK108 pKa = 10.48IEE110 pKa = 4.25SFLPGPKK117 pKa = 9.13GTWNDD122 pKa = 3.15QLYY125 pKa = 10.91SVGLWDD131 pKa = 4.01AAVALVTRR139 pKa = 11.84QSTLDD144 pKa = 3.53EE145 pKa = 4.93LGLRR149 pKa = 11.84TPTLEE154 pKa = 4.19EE155 pKa = 4.0PWTLEE160 pKa = 3.76EE161 pKa = 5.23FMGAMEE167 pKa = 4.41AAKK170 pKa = 10.64ASGKK174 pKa = 10.05YY175 pKa = 10.04DD176 pKa = 3.5YY177 pKa = 11.18AIDD180 pKa = 5.27LGMSDD185 pKa = 3.01QGEE188 pKa = 4.35WYY190 pKa = 9.03PYY192 pKa = 11.22AFLPFLWSFGGDD204 pKa = 3.37LVDD207 pKa = 4.5VQADD211 pKa = 3.6PQTAEE216 pKa = 4.01GVLNGDD222 pKa = 3.69AGLKK226 pKa = 9.89FGEE229 pKa = 4.02WWQGMFANGYY239 pKa = 10.37APGPSQDD246 pKa = 4.13PADD249 pKa = 4.04HH250 pKa = 7.12ASGFQEE256 pKa = 3.99GKK258 pKa = 10.64YY259 pKa = 10.04AFSWNGNWLALPFLEE274 pKa = 6.02AYY276 pKa = 10.29DD277 pKa = 4.14DD278 pKa = 4.38VVFLPAPDD286 pKa = 4.4FGNGNYY292 pKa = 9.6IGGASWQFAVSGTTEE307 pKa = 3.99HH308 pKa = 7.35PEE310 pKa = 3.73GGAAFIEE317 pKa = 4.86FALQDD322 pKa = 3.24KK323 pKa = 10.5YY324 pKa = 11.16LAQFSDD330 pKa = 5.05GIGLIPPTTTAAAMTQNYY348 pKa = 9.13KK349 pKa = 10.57EE350 pKa = 4.35SGPMAVFYY358 pKa = 10.81DD359 pKa = 4.38LSADD363 pKa = 3.67QARR366 pKa = 11.84LRR368 pKa = 11.84PVTPGYY374 pKa = 9.65PVASKK379 pKa = 11.12VFLKK383 pKa = 11.21AMMDD387 pKa = 3.45IANGADD393 pKa = 3.34VADD396 pKa = 4.32TLDD399 pKa = 3.68SAVDD403 pKa = 4.27EE404 pKa = 4.58IDD406 pKa = 5.5ADD408 pKa = 3.53IQANNGYY415 pKa = 9.69AQAQQ419 pKa = 3.46
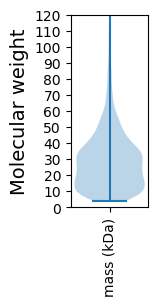
Molecular weight: 45.2 kDa
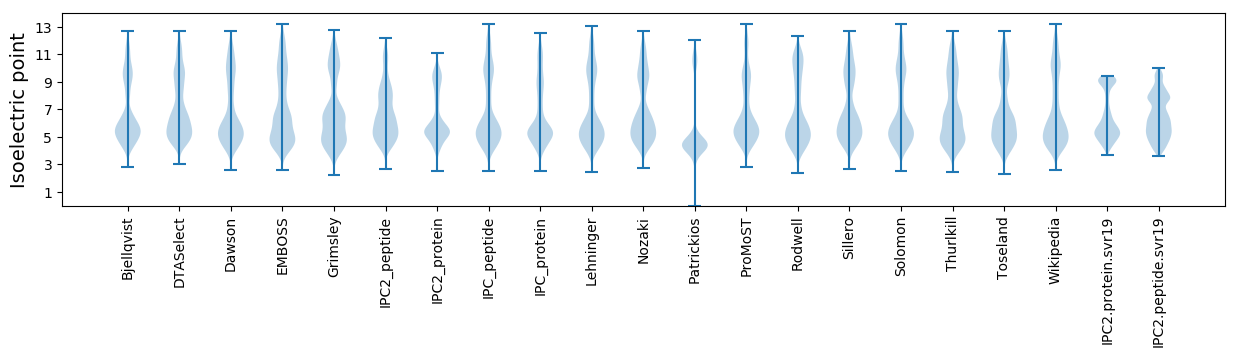
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A017HQN9|A0A017HQN9_9RHOB Recombinase OS=Rubellimicrobium mesophilum DSM 19309 OX=442562 GN=Rumeso_02468 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.41VLCAA44 pKa = 3.88
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 9.2ARR15 pKa = 11.84HH16 pKa = 4.71GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84AVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.41VLCAA44 pKa = 3.88
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1389346 |
37 |
2011 |
280.9 |
30.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.996 ± 0.051 | 0.818 ± 0.011 |
5.774 ± 0.033 | 6.122 ± 0.035 |
3.29 ± 0.02 | 9.43 ± 0.047 |
2.095 ± 0.018 | 4.135 ± 0.026 |
2.2 ± 0.025 | 10.56 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.018 | 2.084 ± 0.018 |
5.83 ± 0.033 | 2.99 ± 0.023 |
8.19 ± 0.044 | 4.881 ± 0.027 |
5.194 ± 0.028 | 7.527 ± 0.026 |
1.563 ± 0.016 | 1.916 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
