
Erythrobacter sp. YT30
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Erythrobacter/Porphyrobacter group; Erythrobacter; unclassified Erythrobacter
Average proteome isoelectric point is 5.78
Get precalculated fractions of proteins
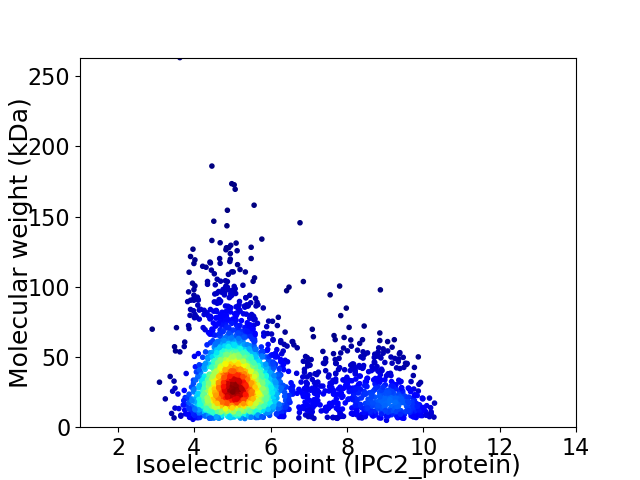
Virtual 2D-PAGE plot for 2997 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A109LNU1|A0A109LNU1_9SPHN Uncharacterized protein OS=Erythrobacter sp. YT30 OX=1735012 GN=AUC45_07155 PE=3 SV=1
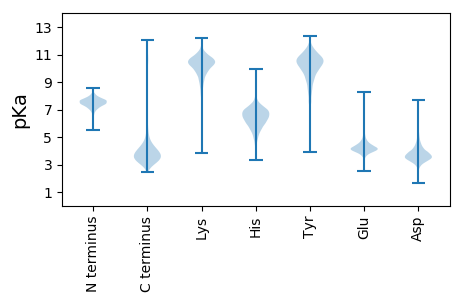
MM1 pKa = 7.21TAILDD6 pKa = 3.81AAGGDD11 pKa = 3.76TAIEE15 pKa = 4.01NSLLEE20 pKa = 4.87ADD22 pKa = 5.03DD23 pKa = 4.62EE24 pKa = 4.79EE25 pKa = 4.83SLQTQILGLTPEE37 pKa = 4.14YY38 pKa = 10.97SGGNFDD44 pKa = 5.5LATSSSRR51 pKa = 11.84LAAAHH56 pKa = 6.88LYY58 pKa = 10.34DD59 pKa = 4.65HH60 pKa = 6.71EE61 pKa = 4.92TMFSVSDD68 pKa = 3.24TRR70 pKa = 11.84IWVEE74 pKa = 3.81SYY76 pKa = 11.1YY77 pKa = 11.24LNGDD81 pKa = 3.66YY82 pKa = 11.09DD83 pKa = 4.3PDD85 pKa = 5.58SIAGGYY91 pKa = 9.56SLGSFGLTGGYY102 pKa = 8.73EE103 pKa = 3.79AGTPFGRR110 pKa = 11.84VGLSANISWGSNSNGTVDD128 pKa = 3.88AEE130 pKa = 4.31VDD132 pKa = 3.29ASHH135 pKa = 6.96YY136 pKa = 9.4EE137 pKa = 4.01AALHH141 pKa = 5.78WRR143 pKa = 11.84EE144 pKa = 3.98QLGGLLMFARR154 pKa = 11.84GSFAYY159 pKa = 9.97VGYY162 pKa = 10.04SSEE165 pKa = 4.1RR166 pKa = 11.84TFEE169 pKa = 4.26GDD171 pKa = 4.03LDD173 pKa = 3.92GTADD177 pKa = 4.83DD178 pKa = 6.39DD179 pKa = 5.19EE180 pKa = 7.72DD181 pKa = 5.7DD182 pKa = 5.2DD183 pKa = 4.07EE184 pKa = 7.14AGFLRR189 pKa = 11.84VASGDD194 pKa = 3.66FDD196 pKa = 3.82GLLYY200 pKa = 10.81SGLVGASYY208 pKa = 9.88QAKK211 pKa = 9.68LGQRR215 pKa = 11.84FTLTPLVSLEE225 pKa = 3.81YY226 pKa = 10.57FRR228 pKa = 11.84LDD230 pKa = 3.08EE231 pKa = 5.51DD232 pKa = 4.69GYY234 pKa = 11.04EE235 pKa = 4.11EE236 pKa = 4.25TGGGDD241 pKa = 3.52GMNLIVEE248 pKa = 4.6DD249 pKa = 4.54RR250 pKa = 11.84SSDD253 pKa = 3.65SLSINSTLALAYY265 pKa = 10.1AIGPQQPDD273 pKa = 4.5MIPMSIEE280 pKa = 3.38IEE282 pKa = 3.92AGRR285 pKa = 11.84RR286 pKa = 11.84TVLNGSLGDD295 pKa = 4.07TIAEE299 pKa = 4.15FEE301 pKa = 4.39DD302 pKa = 3.69VGDD305 pKa = 4.31SFRR308 pKa = 11.84LMAPDD313 pKa = 5.47IEE315 pKa = 4.64SAWLGEE321 pKa = 3.64ISIIGGGYY329 pKa = 10.27DD330 pKa = 3.86FLWSVNAAAEE340 pKa = 4.34HH341 pKa = 6.01EE342 pKa = 4.45GDD344 pKa = 3.29RR345 pKa = 11.84TVYY348 pKa = 10.32SLRR351 pKa = 11.84AGLGIAFF358 pKa = 4.81
MM1 pKa = 7.21TAILDD6 pKa = 3.81AAGGDD11 pKa = 3.76TAIEE15 pKa = 4.01NSLLEE20 pKa = 4.87ADD22 pKa = 5.03DD23 pKa = 4.62EE24 pKa = 4.79EE25 pKa = 4.83SLQTQILGLTPEE37 pKa = 4.14YY38 pKa = 10.97SGGNFDD44 pKa = 5.5LATSSSRR51 pKa = 11.84LAAAHH56 pKa = 6.88LYY58 pKa = 10.34DD59 pKa = 4.65HH60 pKa = 6.71EE61 pKa = 4.92TMFSVSDD68 pKa = 3.24TRR70 pKa = 11.84IWVEE74 pKa = 3.81SYY76 pKa = 11.1YY77 pKa = 11.24LNGDD81 pKa = 3.66YY82 pKa = 11.09DD83 pKa = 4.3PDD85 pKa = 5.58SIAGGYY91 pKa = 9.56SLGSFGLTGGYY102 pKa = 8.73EE103 pKa = 3.79AGTPFGRR110 pKa = 11.84VGLSANISWGSNSNGTVDD128 pKa = 3.88AEE130 pKa = 4.31VDD132 pKa = 3.29ASHH135 pKa = 6.96YY136 pKa = 9.4EE137 pKa = 4.01AALHH141 pKa = 5.78WRR143 pKa = 11.84EE144 pKa = 3.98QLGGLLMFARR154 pKa = 11.84GSFAYY159 pKa = 9.97VGYY162 pKa = 10.04SSEE165 pKa = 4.1RR166 pKa = 11.84TFEE169 pKa = 4.26GDD171 pKa = 4.03LDD173 pKa = 3.92GTADD177 pKa = 4.83DD178 pKa = 6.39DD179 pKa = 5.19EE180 pKa = 7.72DD181 pKa = 5.7DD182 pKa = 5.2DD183 pKa = 4.07EE184 pKa = 7.14AGFLRR189 pKa = 11.84VASGDD194 pKa = 3.66FDD196 pKa = 3.82GLLYY200 pKa = 10.81SGLVGASYY208 pKa = 9.88QAKK211 pKa = 9.68LGQRR215 pKa = 11.84FTLTPLVSLEE225 pKa = 3.81YY226 pKa = 10.57FRR228 pKa = 11.84LDD230 pKa = 3.08EE231 pKa = 5.51DD232 pKa = 4.69GYY234 pKa = 11.04EE235 pKa = 4.11EE236 pKa = 4.25TGGGDD241 pKa = 3.52GMNLIVEE248 pKa = 4.6DD249 pKa = 4.54RR250 pKa = 11.84SSDD253 pKa = 3.65SLSINSTLALAYY265 pKa = 10.1AIGPQQPDD273 pKa = 4.5MIPMSIEE280 pKa = 3.38IEE282 pKa = 3.92AGRR285 pKa = 11.84RR286 pKa = 11.84TVLNGSLGDD295 pKa = 4.07TIAEE299 pKa = 4.15FEE301 pKa = 4.39DD302 pKa = 3.69VGDD305 pKa = 4.31SFRR308 pKa = 11.84LMAPDD313 pKa = 5.47IEE315 pKa = 4.64SAWLGEE321 pKa = 3.64ISIIGGGYY329 pKa = 10.27DD330 pKa = 3.86FLWSVNAAAEE340 pKa = 4.34HH341 pKa = 6.01EE342 pKa = 4.45GDD344 pKa = 3.29RR345 pKa = 11.84TVYY348 pKa = 10.32SLRR351 pKa = 11.84AGLGIAFF358 pKa = 4.81
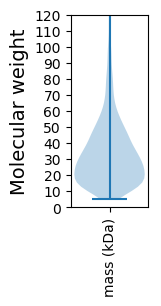
Molecular weight: 38.24 kDa
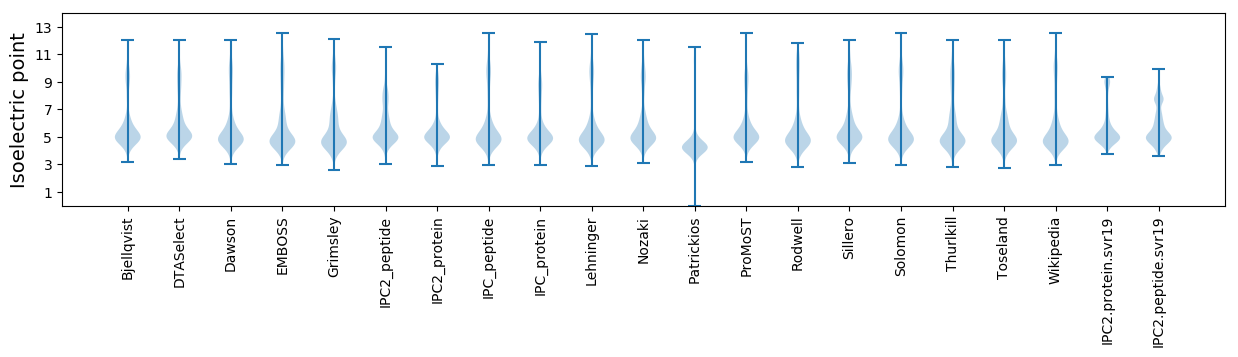
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A109LT76|A0A109LT76_9SPHN Uncharacterized protein OS=Erythrobacter sp. YT30 OX=1735012 GN=AUC45_04365 PE=4 SV=1
MM1 pKa = 6.85NTNLKK6 pKa = 10.4LFLSALTLAFVSTPVVAEE24 pKa = 3.81QVTVSVEE31 pKa = 3.99HH32 pKa = 7.39ADD34 pKa = 3.98LNLSNEE40 pKa = 4.17KK41 pKa = 10.69GIEE44 pKa = 4.11TLKK47 pKa = 10.83GRR49 pKa = 11.84LNGAINRR56 pKa = 11.84VCGPVGFRR64 pKa = 11.84SVTQIYY70 pKa = 9.54RR71 pKa = 11.84ARR73 pKa = 11.84KK74 pKa = 8.29CRR76 pKa = 11.84RR77 pKa = 11.84TLRR80 pKa = 11.84RR81 pKa = 11.84QARR84 pKa = 11.84MLLSEE89 pKa = 3.92IRR91 pKa = 11.84GGNVAIASNFEE102 pKa = 3.42IEE104 pKa = 4.21YY105 pKa = 10.56RR106 pKa = 11.84GG107 pKa = 3.37
MM1 pKa = 6.85NTNLKK6 pKa = 10.4LFLSALTLAFVSTPVVAEE24 pKa = 3.81QVTVSVEE31 pKa = 3.99HH32 pKa = 7.39ADD34 pKa = 3.98LNLSNEE40 pKa = 4.17KK41 pKa = 10.69GIEE44 pKa = 4.11TLKK47 pKa = 10.83GRR49 pKa = 11.84LNGAINRR56 pKa = 11.84VCGPVGFRR64 pKa = 11.84SVTQIYY70 pKa = 9.54RR71 pKa = 11.84ARR73 pKa = 11.84KK74 pKa = 8.29CRR76 pKa = 11.84RR77 pKa = 11.84TLRR80 pKa = 11.84RR81 pKa = 11.84QARR84 pKa = 11.84MLLSEE89 pKa = 3.92IRR91 pKa = 11.84GGNVAIASNFEE102 pKa = 3.42IEE104 pKa = 4.21YY105 pKa = 10.56RR106 pKa = 11.84GG107 pKa = 3.37
Molecular weight: 11.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
959551 |
42 |
2579 |
320.2 |
34.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.926 ± 0.053 | 0.818 ± 0.014 |
6.296 ± 0.034 | 6.731 ± 0.043 |
3.88 ± 0.029 | 8.531 ± 0.046 |
1.852 ± 0.021 | 5.431 ± 0.027 |
3.548 ± 0.037 | 9.456 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.51 ± 0.026 | 2.946 ± 0.032 |
4.939 ± 0.033 | 3.226 ± 0.025 |
6.408 ± 0.047 | 5.789 ± 0.032 |
5.31 ± 0.036 | 6.812 ± 0.037 |
1.369 ± 0.02 | 2.222 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
