
Qipengyuania marisflavi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Qipengyuania
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
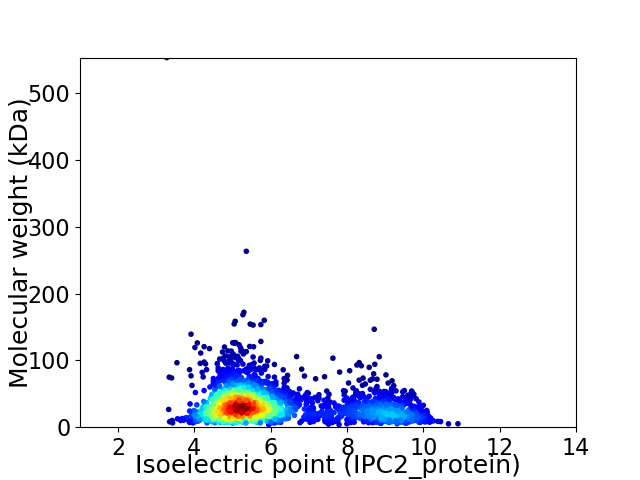
Virtual 2D-PAGE plot for 2508 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S3P2T8|A0A5S3P2T8_9SPHN S9 family peptidase OS=Qipengyuania marisflavi OX=2486356 GN=FEV51_10385 PE=4 SV=1
MM1 pKa = 7.78SYY3 pKa = 11.05LFDD6 pKa = 3.33QRR8 pKa = 11.84SFQFNRR14 pKa = 11.84FDD16 pKa = 5.23ADD18 pKa = 2.95MGLNFGRR25 pKa = 11.84AGSRR29 pKa = 11.84AAMWSDD35 pKa = 3.33VGVSHH40 pKa = 7.69HH41 pKa = 7.47DD42 pKa = 3.33GHH44 pKa = 7.65DD45 pKa = 3.6HH46 pKa = 7.28FKK48 pKa = 8.34MTASLFADD56 pKa = 3.86STSSQAPVDD65 pKa = 4.08LAADD69 pKa = 3.76VPGDD73 pKa = 3.42SGTGVTVTIGDD84 pKa = 3.7MVYY87 pKa = 11.46DD88 pKa = 3.94MLEE91 pKa = 3.93VAGDD95 pKa = 3.94HH96 pKa = 6.56DD97 pKa = 4.08WFRR100 pKa = 11.84IDD102 pKa = 4.1LAAGQTLSIVLTGYY116 pKa = 11.23GDD118 pKa = 4.35DD119 pKa = 4.47SLIDD123 pKa = 3.85PYY125 pKa = 11.4LYY127 pKa = 10.95LRR129 pKa = 11.84DD130 pKa = 3.55ASGNLVAEE138 pKa = 4.7NDD140 pKa = 3.93DD141 pKa = 3.71VTPGSDD147 pKa = 2.78RR148 pKa = 11.84NSRR151 pKa = 11.84IDD153 pKa = 4.26FEE155 pKa = 4.36ATTAGTYY162 pKa = 10.51YY163 pKa = 10.05IDD165 pKa = 3.2VGAWDD170 pKa = 3.74EE171 pKa = 4.99GYY173 pKa = 11.1AGDD176 pKa = 3.93YY177 pKa = 9.83QLQIFDD183 pKa = 4.72SEE185 pKa = 4.42ATTDD189 pKa = 4.32AITANASTTASMLVGAPFHH208 pKa = 6.53GVIEE212 pKa = 4.54TPSDD216 pKa = 3.78HH217 pKa = 7.32DD218 pKa = 3.5WVAIEE223 pKa = 4.55LVAGTEE229 pKa = 4.19YY230 pKa = 10.84QVDD233 pKa = 4.07LSGAGPNALNNPYY246 pKa = 10.55LIVRR250 pKa = 11.84DD251 pKa = 3.44AAGNIVAEE259 pKa = 4.16NDD261 pKa = 3.28NTNGLDD267 pKa = 3.53SAVTFTATTSGTYY280 pKa = 10.62YY281 pKa = 10.0IDD283 pKa = 3.53ASAAFSGGTGEE294 pKa = 4.16YY295 pKa = 9.7LIEE298 pKa = 4.24VTPPLPIFTFDD309 pKa = 3.91QIANQLTNTFWGGEE323 pKa = 3.61QHH325 pKa = 6.72AFVVGADD332 pKa = 3.4NSLTVNLTGINADD345 pKa = 3.57GQGLARR351 pKa = 11.84AALQSWTDD359 pKa = 3.13VTGIQFVEE367 pKa = 4.3TNGAAEE373 pKa = 4.21ITFQDD378 pKa = 3.86HH379 pKa = 6.22EE380 pKa = 4.81SGAFASSTYY389 pKa = 9.71NGSGAIVSSTVNVSTQWIANYY410 pKa = 7.94GTALDD415 pKa = 4.62DD416 pKa = 3.68YY417 pKa = 11.06SYY419 pKa = 10.16QTFIHH424 pKa = 6.78EE425 pKa = 4.35IGHH428 pKa = 6.36ALGLGHH434 pKa = 7.13GGNYY438 pKa = 9.74NGSAEE443 pKa = 4.26YY444 pKa = 10.01PDD446 pKa = 4.38DD447 pKa = 3.5ATYY450 pKa = 11.25QNDD453 pKa = 3.06SWATTVMSYY462 pKa = 10.2FSQTEE467 pKa = 3.36NDD469 pKa = 3.5YY470 pKa = 11.12FADD473 pKa = 3.42QGFTRR478 pKa = 11.84ASVTTPMNADD488 pKa = 3.24VVAMRR493 pKa = 11.84VLYY496 pKa = 10.47GLSSDD501 pKa = 3.39TRR503 pKa = 11.84LGNTTYY509 pKa = 11.15GFNSNAGSTAFDD521 pKa = 2.97ATVFNDD527 pKa = 3.49TAYY530 pKa = 9.9TIVDD534 pKa = 3.41SGGIDD539 pKa = 3.24TLDD542 pKa = 3.17YY543 pKa = 11.52SGFSQDD549 pKa = 3.26QLIDD553 pKa = 3.62LRR555 pKa = 11.84PEE557 pKa = 3.62YY558 pKa = 10.83FMNIGPAIGNVMIGRR573 pKa = 11.84GTFIEE578 pKa = 4.14NAIGGSGADD587 pKa = 4.07TINGNQFNNALTGWAGADD605 pKa = 3.58VISGFGGRR613 pKa = 11.84DD614 pKa = 3.18TLQGGNGNDD623 pKa = 3.64TLNGGQHH630 pKa = 7.12LDD632 pKa = 3.37TVYY635 pKa = 11.2GGGGADD641 pKa = 4.27AINGGGANDD650 pKa = 4.08LLYY653 pKa = 11.31GEE655 pKa = 5.41GGNDD659 pKa = 3.7TILGANGNDD668 pKa = 4.08YY669 pKa = 11.07IDD671 pKa = 4.41GGAGNDD677 pKa = 4.01TINGGVGSDD686 pKa = 4.02EE687 pKa = 4.19IFGGDD692 pKa = 3.67GDD694 pKa = 4.1DD695 pKa = 4.48TISGGHH701 pKa = 6.13GLDD704 pKa = 4.07VIHH707 pKa = 7.09GGLGEE712 pKa = 4.01DD713 pKa = 4.42TINGGSFADD722 pKa = 4.42TIFGDD727 pKa = 3.87EE728 pKa = 4.78GNDD731 pKa = 3.57TLLGGPGSDD740 pKa = 3.83TIYY743 pKa = 11.11GGADD747 pKa = 3.19NDD749 pKa = 4.74SISGGNANDD758 pKa = 3.76TLYY761 pKa = 11.44GDD763 pKa = 4.87GGNDD767 pKa = 3.71TINGNNGADD776 pKa = 4.0TIDD779 pKa = 3.73GGAGIDD785 pKa = 3.79RR786 pKa = 11.84ISGGLAADD794 pKa = 3.71ILTGGGGNDD803 pKa = 3.29LFVFDD808 pKa = 5.1SLLGGPNIDD817 pKa = 4.19QITDD821 pKa = 3.75FTSGSDD827 pKa = 3.64TINLDD832 pKa = 3.34RR833 pKa = 11.84AIFTGIGSDD842 pKa = 3.53GALAGSAFRR851 pKa = 11.84VGTAAQDD858 pKa = 3.27ADD860 pKa = 3.69DD861 pKa = 5.11RR862 pKa = 11.84IIYY865 pKa = 7.89TRR867 pKa = 11.84STGEE871 pKa = 3.45IFYY874 pKa = 10.88DD875 pKa = 3.57ADD877 pKa = 3.83GVGGADD883 pKa = 2.64AVLFAKK889 pKa = 10.29VDD891 pKa = 3.33AGMVLSASDD900 pKa = 3.81FAAFSASAAAGSGAKK915 pKa = 10.46GGDD918 pKa = 3.2AMFAPLEE925 pKa = 4.25SLHH928 pKa = 6.54GVDD931 pKa = 4.53IFAIVV936 pKa = 2.83
MM1 pKa = 7.78SYY3 pKa = 11.05LFDD6 pKa = 3.33QRR8 pKa = 11.84SFQFNRR14 pKa = 11.84FDD16 pKa = 5.23ADD18 pKa = 2.95MGLNFGRR25 pKa = 11.84AGSRR29 pKa = 11.84AAMWSDD35 pKa = 3.33VGVSHH40 pKa = 7.69HH41 pKa = 7.47DD42 pKa = 3.33GHH44 pKa = 7.65DD45 pKa = 3.6HH46 pKa = 7.28FKK48 pKa = 8.34MTASLFADD56 pKa = 3.86STSSQAPVDD65 pKa = 4.08LAADD69 pKa = 3.76VPGDD73 pKa = 3.42SGTGVTVTIGDD84 pKa = 3.7MVYY87 pKa = 11.46DD88 pKa = 3.94MLEE91 pKa = 3.93VAGDD95 pKa = 3.94HH96 pKa = 6.56DD97 pKa = 4.08WFRR100 pKa = 11.84IDD102 pKa = 4.1LAAGQTLSIVLTGYY116 pKa = 11.23GDD118 pKa = 4.35DD119 pKa = 4.47SLIDD123 pKa = 3.85PYY125 pKa = 11.4LYY127 pKa = 10.95LRR129 pKa = 11.84DD130 pKa = 3.55ASGNLVAEE138 pKa = 4.7NDD140 pKa = 3.93DD141 pKa = 3.71VTPGSDD147 pKa = 2.78RR148 pKa = 11.84NSRR151 pKa = 11.84IDD153 pKa = 4.26FEE155 pKa = 4.36ATTAGTYY162 pKa = 10.51YY163 pKa = 10.05IDD165 pKa = 3.2VGAWDD170 pKa = 3.74EE171 pKa = 4.99GYY173 pKa = 11.1AGDD176 pKa = 3.93YY177 pKa = 9.83QLQIFDD183 pKa = 4.72SEE185 pKa = 4.42ATTDD189 pKa = 4.32AITANASTTASMLVGAPFHH208 pKa = 6.53GVIEE212 pKa = 4.54TPSDD216 pKa = 3.78HH217 pKa = 7.32DD218 pKa = 3.5WVAIEE223 pKa = 4.55LVAGTEE229 pKa = 4.19YY230 pKa = 10.84QVDD233 pKa = 4.07LSGAGPNALNNPYY246 pKa = 10.55LIVRR250 pKa = 11.84DD251 pKa = 3.44AAGNIVAEE259 pKa = 4.16NDD261 pKa = 3.28NTNGLDD267 pKa = 3.53SAVTFTATTSGTYY280 pKa = 10.62YY281 pKa = 10.0IDD283 pKa = 3.53ASAAFSGGTGEE294 pKa = 4.16YY295 pKa = 9.7LIEE298 pKa = 4.24VTPPLPIFTFDD309 pKa = 3.91QIANQLTNTFWGGEE323 pKa = 3.61QHH325 pKa = 6.72AFVVGADD332 pKa = 3.4NSLTVNLTGINADD345 pKa = 3.57GQGLARR351 pKa = 11.84AALQSWTDD359 pKa = 3.13VTGIQFVEE367 pKa = 4.3TNGAAEE373 pKa = 4.21ITFQDD378 pKa = 3.86HH379 pKa = 6.22EE380 pKa = 4.81SGAFASSTYY389 pKa = 9.71NGSGAIVSSTVNVSTQWIANYY410 pKa = 7.94GTALDD415 pKa = 4.62DD416 pKa = 3.68YY417 pKa = 11.06SYY419 pKa = 10.16QTFIHH424 pKa = 6.78EE425 pKa = 4.35IGHH428 pKa = 6.36ALGLGHH434 pKa = 7.13GGNYY438 pKa = 9.74NGSAEE443 pKa = 4.26YY444 pKa = 10.01PDD446 pKa = 4.38DD447 pKa = 3.5ATYY450 pKa = 11.25QNDD453 pKa = 3.06SWATTVMSYY462 pKa = 10.2FSQTEE467 pKa = 3.36NDD469 pKa = 3.5YY470 pKa = 11.12FADD473 pKa = 3.42QGFTRR478 pKa = 11.84ASVTTPMNADD488 pKa = 3.24VVAMRR493 pKa = 11.84VLYY496 pKa = 10.47GLSSDD501 pKa = 3.39TRR503 pKa = 11.84LGNTTYY509 pKa = 11.15GFNSNAGSTAFDD521 pKa = 2.97ATVFNDD527 pKa = 3.49TAYY530 pKa = 9.9TIVDD534 pKa = 3.41SGGIDD539 pKa = 3.24TLDD542 pKa = 3.17YY543 pKa = 11.52SGFSQDD549 pKa = 3.26QLIDD553 pKa = 3.62LRR555 pKa = 11.84PEE557 pKa = 3.62YY558 pKa = 10.83FMNIGPAIGNVMIGRR573 pKa = 11.84GTFIEE578 pKa = 4.14NAIGGSGADD587 pKa = 4.07TINGNQFNNALTGWAGADD605 pKa = 3.58VISGFGGRR613 pKa = 11.84DD614 pKa = 3.18TLQGGNGNDD623 pKa = 3.64TLNGGQHH630 pKa = 7.12LDD632 pKa = 3.37TVYY635 pKa = 11.2GGGGADD641 pKa = 4.27AINGGGANDD650 pKa = 4.08LLYY653 pKa = 11.31GEE655 pKa = 5.41GGNDD659 pKa = 3.7TILGANGNDD668 pKa = 4.08YY669 pKa = 11.07IDD671 pKa = 4.41GGAGNDD677 pKa = 4.01TINGGVGSDD686 pKa = 4.02EE687 pKa = 4.19IFGGDD692 pKa = 3.67GDD694 pKa = 4.1DD695 pKa = 4.48TISGGHH701 pKa = 6.13GLDD704 pKa = 4.07VIHH707 pKa = 7.09GGLGEE712 pKa = 4.01DD713 pKa = 4.42TINGGSFADD722 pKa = 4.42TIFGDD727 pKa = 3.87EE728 pKa = 4.78GNDD731 pKa = 3.57TLLGGPGSDD740 pKa = 3.83TIYY743 pKa = 11.11GGADD747 pKa = 3.19NDD749 pKa = 4.74SISGGNANDD758 pKa = 3.76TLYY761 pKa = 11.44GDD763 pKa = 4.87GGNDD767 pKa = 3.71TINGNNGADD776 pKa = 4.0TIDD779 pKa = 3.73GGAGIDD785 pKa = 3.79RR786 pKa = 11.84ISGGLAADD794 pKa = 3.71ILTGGGGNDD803 pKa = 3.29LFVFDD808 pKa = 5.1SLLGGPNIDD817 pKa = 4.19QITDD821 pKa = 3.75FTSGSDD827 pKa = 3.64TINLDD832 pKa = 3.34RR833 pKa = 11.84AIFTGIGSDD842 pKa = 3.53GALAGSAFRR851 pKa = 11.84VGTAAQDD858 pKa = 3.27ADD860 pKa = 3.69DD861 pKa = 5.11RR862 pKa = 11.84IIYY865 pKa = 7.89TRR867 pKa = 11.84STGEE871 pKa = 3.45IFYY874 pKa = 10.88DD875 pKa = 3.57ADD877 pKa = 3.83GVGGADD883 pKa = 2.64AVLFAKK889 pKa = 10.29VDD891 pKa = 3.33AGMVLSASDD900 pKa = 3.81FAAFSASAAAGSGAKK915 pKa = 10.46GGDD918 pKa = 3.2AMFAPLEE925 pKa = 4.25SLHH928 pKa = 6.54GVDD931 pKa = 4.53IFAIVV936 pKa = 2.83
Molecular weight: 96.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S3P3F6|A0A5S3P3F6_9SPHN DUF969 domain-containing protein OS=Qipengyuania marisflavi OX=2486356 GN=FEV51_09430 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.89GFFARR21 pKa = 11.84KK22 pKa = 7.42ATPGGRR28 pKa = 11.84KK29 pKa = 7.9VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.78KK41 pKa = 10.52LCAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.89GFFARR21 pKa = 11.84KK22 pKa = 7.42ATPGGRR28 pKa = 11.84KK29 pKa = 7.9VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.78KK41 pKa = 10.52LCAA44 pKa = 3.96
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
813454 |
39 |
5844 |
324.3 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.014 ± 0.082 | 0.825 ± 0.016 |
6.261 ± 0.043 | 5.957 ± 0.056 |
3.617 ± 0.029 | 8.878 ± 0.083 |
1.993 ± 0.027 | 5.009 ± 0.032 |
3.207 ± 0.048 | 9.833 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.587 ± 0.028 | 2.563 ± 0.034 |
5.052 ± 0.042 | 3.304 ± 0.027 |
6.946 ± 0.057 | 5.265 ± 0.044 |
5.212 ± 0.05 | 6.898 ± 0.037 |
1.421 ± 0.025 | 2.159 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
