
Halococcus saccharolyticus DSM 5350
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Halococcaceae; Halococcus; Halococcus saccharolyticus
Average proteome isoelectric point is 4.93
Get precalculated fractions of proteins
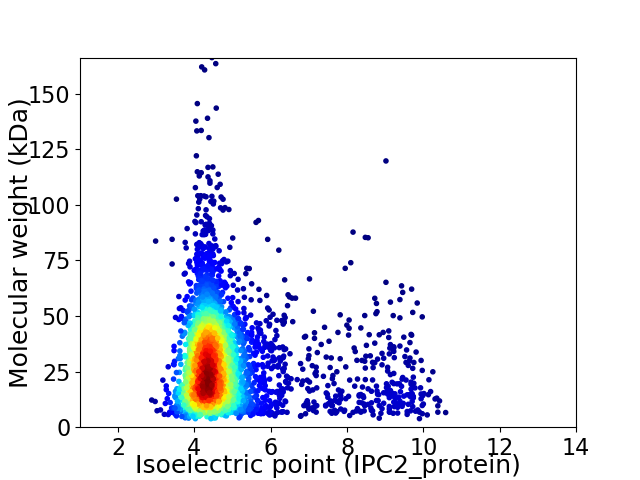
Virtual 2D-PAGE plot for 3441 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
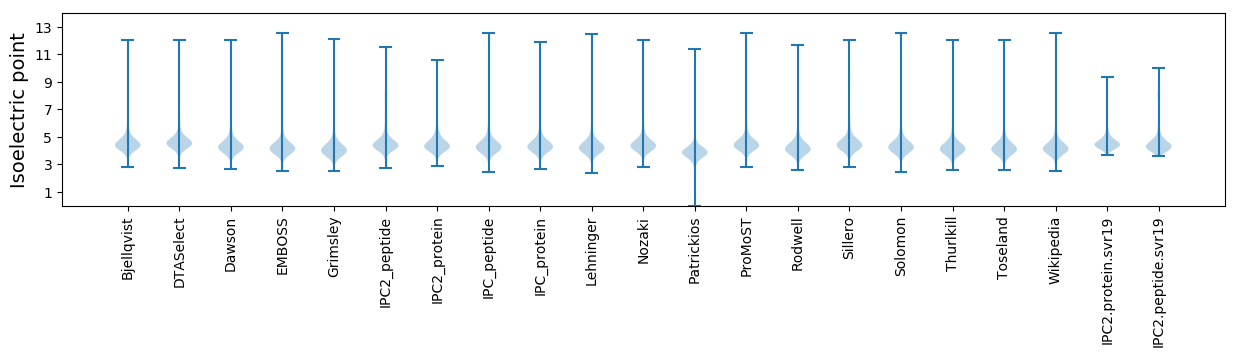
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0MK97|M0MK97_9EURY Uncharacterized protein OS=Halococcus saccharolyticus DSM 5350 OX=1227455 GN=C449_06081 PE=4 SV=1
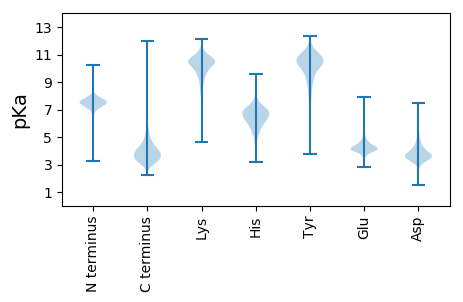
MM1 pKa = 7.67AGLAGCTGGGGGGGNGSGGNEE22 pKa = 4.11SGGGANGSSGNGSGSNGSSANGSSSGGSSGNLTVLHH58 pKa = 6.8GWTGGDD64 pKa = 3.55GATALDD70 pKa = 3.74NLVEE74 pKa = 4.47GFEE77 pKa = 4.21EE78 pKa = 4.42ANPDD82 pKa = 3.05VSANFQAIGGGGNTNLDD99 pKa = 3.57TTISNRR105 pKa = 11.84AQNQNLPASWADD117 pKa = 3.34WPGLNLVPFTEE128 pKa = 4.8AGLLADD134 pKa = 4.12VGEE137 pKa = 4.49QVWTDD142 pKa = 3.7DD143 pKa = 3.73LRR145 pKa = 11.84SNYY148 pKa = 8.87SQEE151 pKa = 3.78AKK153 pKa = 9.86TYY155 pKa = 9.15SQVGEE160 pKa = 4.14DD161 pKa = 3.9AGSIGDD167 pKa = 3.87GPYY170 pKa = 9.66VAVPIGSHH178 pKa = 5.89RR179 pKa = 11.84MNDD182 pKa = 2.92LFYY185 pKa = 10.1NVSVVEE191 pKa = 4.13EE192 pKa = 4.39AGVDD196 pKa = 3.52PTSFSQPSDD205 pKa = 3.3LLAAMKK211 pKa = 9.52TVEE214 pKa = 4.42EE215 pKa = 4.17EE216 pKa = 3.72TDD218 pKa = 3.52AVGMAQGLQAPFTTLQLWEE237 pKa = 4.42VVLQGQAGYY246 pKa = 10.32QPFMDD251 pKa = 4.75YY252 pKa = 11.0INGEE256 pKa = 3.88GDD258 pKa = 3.38EE259 pKa = 4.21QAVRR263 pKa = 11.84SAFEE267 pKa = 3.73MVNQYY272 pKa = 10.42FNHH275 pKa = 6.35INQNASSVGFTQANQLIMNGEE296 pKa = 4.31AAFMHH301 pKa = 5.85QGNWVAGAYY310 pKa = 9.77RR311 pKa = 11.84NQDD314 pKa = 2.94GFEE317 pKa = 4.42YY318 pKa = 10.62EE319 pKa = 4.31SDD321 pKa = 3.35WNNVSFPGTDD331 pKa = 3.11DD332 pKa = 3.18MYY334 pKa = 11.66GLHH337 pKa = 7.04LDD339 pKa = 4.25SFPMAASAADD349 pKa = 3.73SQAAQAWLSYY359 pKa = 10.8VGTPDD364 pKa = 2.95AQVRR368 pKa = 11.84FNQYY372 pKa = 9.69KK373 pKa = 10.16GSIPPRR379 pKa = 11.84TDD381 pKa = 2.59VPTDD385 pKa = 3.15EE386 pKa = 4.98FGPYY390 pKa = 8.51LTQTIEE396 pKa = 4.25DD397 pKa = 4.17YY398 pKa = 11.66NNVSNKK404 pKa = 9.65PPTIAHH410 pKa = 6.75GLAVLPDD417 pKa = 3.5VHH419 pKa = 8.25SEE421 pKa = 3.49IDD423 pKa = 3.76GVITDD428 pKa = 4.31SFLGSEE434 pKa = 4.75DD435 pKa = 4.71LDD437 pKa = 4.06AATQGMLDD445 pKa = 3.81AVSGSNN451 pKa = 3.57
MM1 pKa = 7.67AGLAGCTGGGGGGGNGSGGNEE22 pKa = 4.11SGGGANGSSGNGSGSNGSSANGSSSGGSSGNLTVLHH58 pKa = 6.8GWTGGDD64 pKa = 3.55GATALDD70 pKa = 3.74NLVEE74 pKa = 4.47GFEE77 pKa = 4.21EE78 pKa = 4.42ANPDD82 pKa = 3.05VSANFQAIGGGGNTNLDD99 pKa = 3.57TTISNRR105 pKa = 11.84AQNQNLPASWADD117 pKa = 3.34WPGLNLVPFTEE128 pKa = 4.8AGLLADD134 pKa = 4.12VGEE137 pKa = 4.49QVWTDD142 pKa = 3.7DD143 pKa = 3.73LRR145 pKa = 11.84SNYY148 pKa = 8.87SQEE151 pKa = 3.78AKK153 pKa = 9.86TYY155 pKa = 9.15SQVGEE160 pKa = 4.14DD161 pKa = 3.9AGSIGDD167 pKa = 3.87GPYY170 pKa = 9.66VAVPIGSHH178 pKa = 5.89RR179 pKa = 11.84MNDD182 pKa = 2.92LFYY185 pKa = 10.1NVSVVEE191 pKa = 4.13EE192 pKa = 4.39AGVDD196 pKa = 3.52PTSFSQPSDD205 pKa = 3.3LLAAMKK211 pKa = 9.52TVEE214 pKa = 4.42EE215 pKa = 4.17EE216 pKa = 3.72TDD218 pKa = 3.52AVGMAQGLQAPFTTLQLWEE237 pKa = 4.42VVLQGQAGYY246 pKa = 10.32QPFMDD251 pKa = 4.75YY252 pKa = 11.0INGEE256 pKa = 3.88GDD258 pKa = 3.38EE259 pKa = 4.21QAVRR263 pKa = 11.84SAFEE267 pKa = 3.73MVNQYY272 pKa = 10.42FNHH275 pKa = 6.35INQNASSVGFTQANQLIMNGEE296 pKa = 4.31AAFMHH301 pKa = 5.85QGNWVAGAYY310 pKa = 9.77RR311 pKa = 11.84NQDD314 pKa = 2.94GFEE317 pKa = 4.42YY318 pKa = 10.62EE319 pKa = 4.31SDD321 pKa = 3.35WNNVSFPGTDD331 pKa = 3.11DD332 pKa = 3.18MYY334 pKa = 11.66GLHH337 pKa = 7.04LDD339 pKa = 4.25SFPMAASAADD349 pKa = 3.73SQAAQAWLSYY359 pKa = 10.8VGTPDD364 pKa = 2.95AQVRR368 pKa = 11.84FNQYY372 pKa = 9.69KK373 pKa = 10.16GSIPPRR379 pKa = 11.84TDD381 pKa = 2.59VPTDD385 pKa = 3.15EE386 pKa = 4.98FGPYY390 pKa = 8.51LTQTIEE396 pKa = 4.25DD397 pKa = 4.17YY398 pKa = 11.66NNVSNKK404 pKa = 9.65PPTIAHH410 pKa = 6.75GLAVLPDD417 pKa = 3.5VHH419 pKa = 8.25SEE421 pKa = 3.49IDD423 pKa = 3.76GVITDD428 pKa = 4.31SFLGSEE434 pKa = 4.75DD435 pKa = 4.71LDD437 pKa = 4.06AATQGMLDD445 pKa = 3.81AVSGSNN451 pKa = 3.57
Molecular weight: 46.96 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0MLV4|M0MLV4_9EURY HTH_11 domain-containing protein OS=Halococcus saccharolyticus DSM 5350 OX=1227455 GN=C449_03336 PE=4 SV=1
MM1 pKa = 7.53SLPTRR6 pKa = 11.84TNSSLVNTPVRR17 pKa = 11.84FGTTIRR23 pKa = 11.84TEE25 pKa = 4.25DD26 pKa = 3.51EE27 pKa = 3.99VGDD30 pKa = 3.78ARR32 pKa = 11.84SVEE35 pKa = 4.18EE36 pKa = 3.56QVGFVGRR43 pKa = 11.84DD44 pKa = 3.19RR45 pKa = 11.84SATSLRR51 pKa = 11.84DD52 pKa = 3.49ATSEE56 pKa = 3.96YY57 pKa = 11.25AKK59 pKa = 10.68NNKK62 pKa = 9.03AALRR66 pKa = 11.84RR67 pKa = 11.84GFRR70 pKa = 11.84KK71 pKa = 10.38DD72 pKa = 3.04GFVATTARR80 pKa = 11.84SGRR83 pKa = 11.84AVWRR87 pKa = 11.84RR88 pKa = 11.84NEE90 pKa = 4.32SVTDD94 pKa = 3.61TNAA97 pKa = 2.89
MM1 pKa = 7.53SLPTRR6 pKa = 11.84TNSSLVNTPVRR17 pKa = 11.84FGTTIRR23 pKa = 11.84TEE25 pKa = 4.25DD26 pKa = 3.51EE27 pKa = 3.99VGDD30 pKa = 3.78ARR32 pKa = 11.84SVEE35 pKa = 4.18EE36 pKa = 3.56QVGFVGRR43 pKa = 11.84DD44 pKa = 3.19RR45 pKa = 11.84SATSLRR51 pKa = 11.84DD52 pKa = 3.49ATSEE56 pKa = 3.96YY57 pKa = 11.25AKK59 pKa = 10.68NNKK62 pKa = 9.03AALRR66 pKa = 11.84RR67 pKa = 11.84GFRR70 pKa = 11.84KK71 pKa = 10.38DD72 pKa = 3.04GFVATTARR80 pKa = 11.84SGRR83 pKa = 11.84AVWRR87 pKa = 11.84RR88 pKa = 11.84NEE90 pKa = 4.32SVTDD94 pKa = 3.61TNAA97 pKa = 2.89
Molecular weight: 10.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981310 |
34 |
1515 |
285.2 |
30.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.244 ± 0.059 | 0.752 ± 0.012 |
8.22 ± 0.051 | 8.205 ± 0.062 |
3.359 ± 0.027 | 8.862 ± 0.048 |
2.068 ± 0.022 | 4.272 ± 0.031 |
1.691 ± 0.023 | 8.79 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.779 ± 0.016 | 2.472 ± 0.026 |
4.626 ± 0.026 | 2.395 ± 0.023 |
6.559 ± 0.037 | 5.467 ± 0.035 |
6.707 ± 0.035 | 8.764 ± 0.046 |
1.15 ± 0.015 | 2.618 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
