
Poinsettia latent virus (isolate Euphorbia pulcherrima/Germany/Siepen/2005) (PnLV) (Poinsettia cryptic virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polemovirus; Poinsettia latent virus
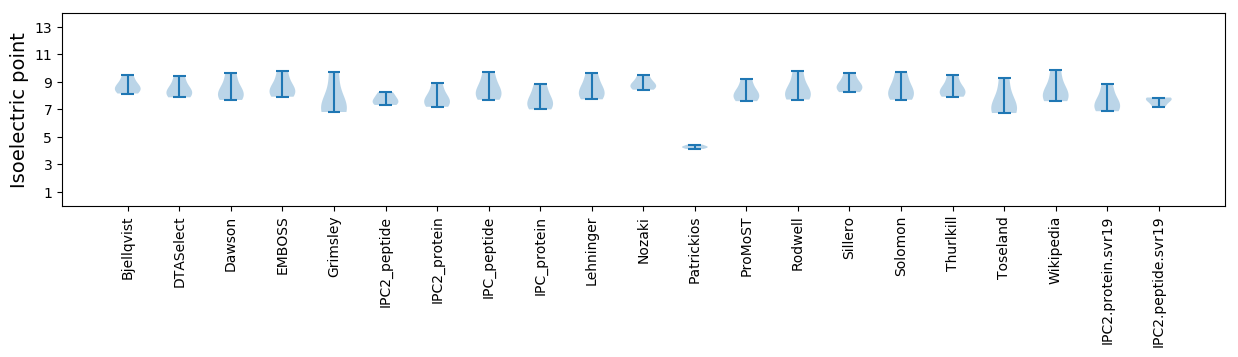
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
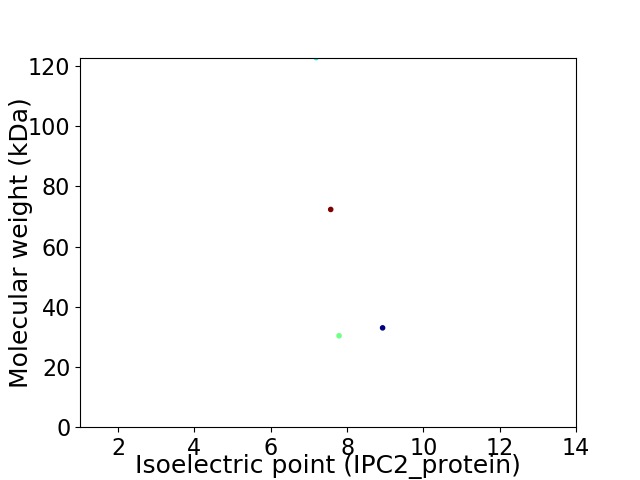
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q5NDN0|P1_PNLV Genome-linked protein precursor OS=Poinsettia latent virus (isolate Euphorbia pulcherrima/Germany/Siepen/2005) OX=686943 GN=ORF2 PE=3 SV=1
MM1 pKa = 7.61ALLGIKK7 pKa = 10.59LMTLVFAAWLSCCHH21 pKa = 6.58SSSALPSSGLSGPCLNHH38 pKa = 6.35SCLLRR43 pKa = 11.84NSLNGASQWGTILHH57 pKa = 6.05SPAVGSNCPPCPMMSIMGCSPPKK80 pKa = 9.61PLQSNSYY87 pKa = 10.36GVLCSTIASKK97 pKa = 10.91AKK99 pKa = 9.96QDD101 pKa = 5.61LKK103 pKa = 11.19LCWKK107 pKa = 9.5EE108 pKa = 3.66VQTRR112 pKa = 11.84SEE114 pKa = 4.34MYY116 pKa = 10.04SKK118 pKa = 10.61RR119 pKa = 11.84ISAALIDD126 pKa = 4.31SLHH129 pKa = 5.83QAVGMLLMIIIWIWSSIFLVVYY151 pKa = 10.12HH152 pKa = 6.09VLAYY156 pKa = 6.93MTTYY160 pKa = 10.05HH161 pKa = 7.06LSSAVCVGFLIFCTICAFRR180 pKa = 11.84LISWICGDD188 pKa = 4.31LLAFNVSGLTPIWVNFSEE206 pKa = 4.71SSCPAGLSLRR216 pKa = 11.84RR217 pKa = 11.84YY218 pKa = 9.1KK219 pKa = 10.7NEE221 pKa = 3.79KK222 pKa = 8.48TVEE225 pKa = 4.27GYY227 pKa = 10.53KK228 pKa = 10.52PFIIPQKK235 pKa = 9.65SPKK238 pKa = 9.83KK239 pKa = 10.38SVIEE243 pKa = 3.9LSFSNGSHH251 pKa = 7.02LGYY254 pKa = 9.18ATCVRR259 pKa = 11.84LWDD262 pKa = 4.04GSICLMTAKK271 pKa = 10.24HH272 pKa = 6.04CLVKK276 pKa = 10.43EE277 pKa = 4.0ALLKK281 pKa = 10.94GRR283 pKa = 11.84VAGHH287 pKa = 5.98SLPVKK292 pKa = 10.7NFDD295 pKa = 5.15LFLTCDD301 pKa = 3.84EE302 pKa = 5.01IDD304 pKa = 3.5FSLLRR309 pKa = 11.84GPKK312 pKa = 8.79QWEE315 pKa = 4.11AYY317 pKa = 10.15LGVKK321 pKa = 9.95GADD324 pKa = 3.58LITSNRR330 pKa = 11.84IGRR333 pKa = 11.84SPVTFYY339 pKa = 11.54NLSKK343 pKa = 10.83DD344 pKa = 4.16GEE346 pKa = 4.29WLANSAQITGRR357 pKa = 11.84HH358 pKa = 5.33GKK360 pKa = 9.48LCSVLSNTSPGDD372 pKa = 3.1SGTPYY377 pKa = 10.45YY378 pKa = 10.45SGKK381 pKa = 10.14NVVGIHH387 pKa = 6.49KK388 pKa = 7.92GTSEE392 pKa = 4.2LEE394 pKa = 4.23NYY396 pKa = 10.8NLMIPIPNIPGLTSPDD412 pKa = 3.41FKK414 pKa = 11.47FEE416 pKa = 4.12TTNVRR421 pKa = 11.84GNLYY425 pKa = 9.87NDD427 pKa = 3.0EE428 pKa = 4.22GFRR431 pKa = 11.84LSVGEE436 pKa = 4.21DD437 pKa = 3.15DD438 pKa = 5.91KK439 pKa = 12.05AEE441 pKa = 3.99HH442 pKa = 5.25WTDD445 pKa = 3.22RR446 pKa = 11.84LMKK449 pKa = 10.54SITFKK454 pKa = 9.45TKK456 pKa = 9.05RR457 pKa = 11.84WADD460 pKa = 3.16WAEE463 pKa = 4.13EE464 pKa = 3.97EE465 pKa = 5.29SEE467 pKa = 4.52SDD469 pKa = 3.59DD470 pKa = 3.46EE471 pKa = 4.6RR472 pKa = 11.84GKK474 pKa = 10.56VVPPAKK480 pKa = 9.57PSNYY484 pKa = 10.11GEE486 pKa = 4.57GCPPEE491 pKa = 4.34HH492 pKa = 6.2NQYY495 pKa = 11.09LSDD498 pKa = 4.4VGDD501 pKa = 4.51LLTKK505 pKa = 10.72VIGPEE510 pKa = 3.93QNEE513 pKa = 4.24KK514 pKa = 10.88CVDD517 pKa = 3.11ILMGIMGVDD526 pKa = 3.43KK527 pKa = 11.6NEE529 pKa = 4.08VAPHH533 pKa = 6.16KK534 pKa = 10.23EE535 pKa = 3.66EE536 pKa = 3.91KK537 pKa = 10.41AEE539 pKa = 3.98KK540 pKa = 9.71GKK542 pKa = 10.59RR543 pKa = 11.84SSGFGHH549 pKa = 6.44GKK551 pKa = 8.45NRR553 pKa = 11.84KK554 pKa = 7.14GTNHH558 pKa = 6.39PMRR561 pKa = 11.84RR562 pKa = 11.84GYY564 pKa = 10.6NFRR567 pKa = 11.84NCKK570 pKa = 9.96KK571 pKa = 10.75GGGQDD576 pKa = 3.01EE577 pKa = 4.91SEE579 pKa = 4.29SHH581 pKa = 6.53RR582 pKa = 11.84EE583 pKa = 3.46ISGRR587 pKa = 11.84DD588 pKa = 3.38PGRR591 pKa = 11.84EE592 pKa = 4.14SNDD595 pKa = 3.04KK596 pKa = 10.79SPQGEE601 pKa = 3.89AEE603 pKa = 3.75EE604 pKa = 4.24FEE606 pKa = 5.37RR607 pKa = 11.84YY608 pKa = 9.07FSSFYY613 pKa = 10.49SWKK616 pKa = 9.96LHH618 pKa = 6.1NSGEE622 pKa = 4.38ANSGFRR628 pKa = 11.84PCGKK632 pKa = 9.13IPKK635 pKa = 9.66FYY637 pKa = 10.38RR638 pKa = 11.84PRR640 pKa = 11.84KK641 pKa = 8.78RR642 pKa = 11.84RR643 pKa = 11.84VSEE646 pKa = 3.54WGQNLARR653 pKa = 11.84KK654 pKa = 9.15HH655 pKa = 6.01SSLGEE660 pKa = 3.73ITQGFGWPEE669 pKa = 3.48AGAEE673 pKa = 4.04AEE675 pKa = 4.15LRR677 pKa = 11.84SLRR680 pKa = 11.84LQAQRR685 pKa = 11.84WLEE688 pKa = 3.84RR689 pKa = 11.84SKK691 pKa = 11.51SSVIPSAIEE700 pKa = 3.64RR701 pKa = 11.84EE702 pKa = 4.27IVISRR707 pKa = 11.84LVEE710 pKa = 3.94SYY712 pKa = 10.48KK713 pKa = 9.93ICRR716 pKa = 11.84SEE718 pKa = 5.19APLCSSGSDD727 pKa = 3.62LSWKK731 pKa = 10.44GFLEE735 pKa = 4.19DD736 pKa = 3.46FRR738 pKa = 11.84EE739 pKa = 4.44AVSSLEE745 pKa = 3.78LDD747 pKa = 3.39AGIGVPYY754 pKa = 9.94IGYY757 pKa = 9.7GYY759 pKa = 6.91PTHH762 pKa = 7.0RR763 pKa = 11.84GWVEE767 pKa = 3.72NPRR770 pKa = 11.84LLPVLSRR777 pKa = 11.84LVYY780 pKa = 10.69ARR782 pKa = 11.84LQRR785 pKa = 11.84LATLSVDD792 pKa = 3.11GKK794 pKa = 9.08TPEE797 pKa = 3.92EE798 pKa = 4.07LVRR801 pKa = 11.84DD802 pKa = 3.98GLVDD806 pKa = 3.64PVRR809 pKa = 11.84VFVKK813 pKa = 10.77GEE815 pKa = 3.59PHH817 pKa = 6.19KK818 pKa = 10.54QSKK821 pKa = 10.21LDD823 pKa = 3.38EE824 pKa = 4.27GRR826 pKa = 11.84YY827 pKa = 8.96RR828 pKa = 11.84LIMSVSLIDD837 pKa = 3.51QLVARR842 pKa = 11.84VLFQKK847 pKa = 10.13QNKK850 pKa = 9.68LEE852 pKa = 4.19LLLWRR857 pKa = 11.84SIPSKK862 pKa = 10.52PGFGLSTVEE871 pKa = 3.97QVEE874 pKa = 4.17EE875 pKa = 4.9FIDD878 pKa = 3.42HH879 pKa = 7.41LARR882 pKa = 11.84VVDD885 pKa = 4.2VKK887 pKa = 11.37SDD889 pKa = 3.68DD890 pKa = 4.52LLEE893 pKa = 4.12NWRR896 pKa = 11.84EE897 pKa = 3.86LMVPTDD903 pKa = 3.69CSGFDD908 pKa = 3.34WSVSDD913 pKa = 4.32WMLKK917 pKa = 10.91DD918 pKa = 3.36EE919 pKa = 4.34MEE921 pKa = 4.09VRR923 pKa = 11.84NRR925 pKa = 11.84LTINCNDD932 pKa = 3.23LTRR935 pKa = 11.84RR936 pKa = 11.84LRR938 pKa = 11.84NSWLYY943 pKa = 10.75CLSNSDD949 pKa = 4.24LALSDD954 pKa = 3.77GSLLAQEE961 pKa = 4.54VPGVQKK967 pKa = 10.68SGSYY971 pKa = 8.27NTSSTNSRR979 pKa = 11.84IRR981 pKa = 11.84VMAAYY986 pKa = 9.45FAGASWAVAIGDD998 pKa = 4.3DD999 pKa = 3.91ALEE1002 pKa = 4.98SIDD1005 pKa = 3.67TTLAVYY1011 pKa = 10.38KK1012 pKa = 10.72SLGFKK1017 pKa = 10.76VEE1019 pKa = 3.91VSEE1022 pKa = 5.07DD1023 pKa = 3.72LEE1025 pKa = 4.48FCSHH1029 pKa = 6.22IFKK1032 pKa = 9.77TRR1034 pKa = 11.84SLAIPVNTSKK1044 pKa = 10.24MLYY1047 pKa = 9.31RR1048 pKa = 11.84LIYY1051 pKa = 10.06GYY1053 pKa = 10.57EE1054 pKa = 4.09PEE1056 pKa = 5.02CGNLDD1061 pKa = 3.42VLRR1064 pKa = 11.84NYY1066 pKa = 10.64LCALASVLHH1075 pKa = 6.07EE1076 pKa = 4.51LRR1078 pKa = 11.84HH1079 pKa = 6.23DD1080 pKa = 3.55QDD1082 pKa = 4.09LVQNLSKK1089 pKa = 10.44WLIPDD1094 pKa = 4.33GSQKK1098 pKa = 10.24ISS1100 pKa = 3.08
MM1 pKa = 7.61ALLGIKK7 pKa = 10.59LMTLVFAAWLSCCHH21 pKa = 6.58SSSALPSSGLSGPCLNHH38 pKa = 6.35SCLLRR43 pKa = 11.84NSLNGASQWGTILHH57 pKa = 6.05SPAVGSNCPPCPMMSIMGCSPPKK80 pKa = 9.61PLQSNSYY87 pKa = 10.36GVLCSTIASKK97 pKa = 10.91AKK99 pKa = 9.96QDD101 pKa = 5.61LKK103 pKa = 11.19LCWKK107 pKa = 9.5EE108 pKa = 3.66VQTRR112 pKa = 11.84SEE114 pKa = 4.34MYY116 pKa = 10.04SKK118 pKa = 10.61RR119 pKa = 11.84ISAALIDD126 pKa = 4.31SLHH129 pKa = 5.83QAVGMLLMIIIWIWSSIFLVVYY151 pKa = 10.12HH152 pKa = 6.09VLAYY156 pKa = 6.93MTTYY160 pKa = 10.05HH161 pKa = 7.06LSSAVCVGFLIFCTICAFRR180 pKa = 11.84LISWICGDD188 pKa = 4.31LLAFNVSGLTPIWVNFSEE206 pKa = 4.71SSCPAGLSLRR216 pKa = 11.84RR217 pKa = 11.84YY218 pKa = 9.1KK219 pKa = 10.7NEE221 pKa = 3.79KK222 pKa = 8.48TVEE225 pKa = 4.27GYY227 pKa = 10.53KK228 pKa = 10.52PFIIPQKK235 pKa = 9.65SPKK238 pKa = 9.83KK239 pKa = 10.38SVIEE243 pKa = 3.9LSFSNGSHH251 pKa = 7.02LGYY254 pKa = 9.18ATCVRR259 pKa = 11.84LWDD262 pKa = 4.04GSICLMTAKK271 pKa = 10.24HH272 pKa = 6.04CLVKK276 pKa = 10.43EE277 pKa = 4.0ALLKK281 pKa = 10.94GRR283 pKa = 11.84VAGHH287 pKa = 5.98SLPVKK292 pKa = 10.7NFDD295 pKa = 5.15LFLTCDD301 pKa = 3.84EE302 pKa = 5.01IDD304 pKa = 3.5FSLLRR309 pKa = 11.84GPKK312 pKa = 8.79QWEE315 pKa = 4.11AYY317 pKa = 10.15LGVKK321 pKa = 9.95GADD324 pKa = 3.58LITSNRR330 pKa = 11.84IGRR333 pKa = 11.84SPVTFYY339 pKa = 11.54NLSKK343 pKa = 10.83DD344 pKa = 4.16GEE346 pKa = 4.29WLANSAQITGRR357 pKa = 11.84HH358 pKa = 5.33GKK360 pKa = 9.48LCSVLSNTSPGDD372 pKa = 3.1SGTPYY377 pKa = 10.45YY378 pKa = 10.45SGKK381 pKa = 10.14NVVGIHH387 pKa = 6.49KK388 pKa = 7.92GTSEE392 pKa = 4.2LEE394 pKa = 4.23NYY396 pKa = 10.8NLMIPIPNIPGLTSPDD412 pKa = 3.41FKK414 pKa = 11.47FEE416 pKa = 4.12TTNVRR421 pKa = 11.84GNLYY425 pKa = 9.87NDD427 pKa = 3.0EE428 pKa = 4.22GFRR431 pKa = 11.84LSVGEE436 pKa = 4.21DD437 pKa = 3.15DD438 pKa = 5.91KK439 pKa = 12.05AEE441 pKa = 3.99HH442 pKa = 5.25WTDD445 pKa = 3.22RR446 pKa = 11.84LMKK449 pKa = 10.54SITFKK454 pKa = 9.45TKK456 pKa = 9.05RR457 pKa = 11.84WADD460 pKa = 3.16WAEE463 pKa = 4.13EE464 pKa = 3.97EE465 pKa = 5.29SEE467 pKa = 4.52SDD469 pKa = 3.59DD470 pKa = 3.46EE471 pKa = 4.6RR472 pKa = 11.84GKK474 pKa = 10.56VVPPAKK480 pKa = 9.57PSNYY484 pKa = 10.11GEE486 pKa = 4.57GCPPEE491 pKa = 4.34HH492 pKa = 6.2NQYY495 pKa = 11.09LSDD498 pKa = 4.4VGDD501 pKa = 4.51LLTKK505 pKa = 10.72VIGPEE510 pKa = 3.93QNEE513 pKa = 4.24KK514 pKa = 10.88CVDD517 pKa = 3.11ILMGIMGVDD526 pKa = 3.43KK527 pKa = 11.6NEE529 pKa = 4.08VAPHH533 pKa = 6.16KK534 pKa = 10.23EE535 pKa = 3.66EE536 pKa = 3.91KK537 pKa = 10.41AEE539 pKa = 3.98KK540 pKa = 9.71GKK542 pKa = 10.59RR543 pKa = 11.84SSGFGHH549 pKa = 6.44GKK551 pKa = 8.45NRR553 pKa = 11.84KK554 pKa = 7.14GTNHH558 pKa = 6.39PMRR561 pKa = 11.84RR562 pKa = 11.84GYY564 pKa = 10.6NFRR567 pKa = 11.84NCKK570 pKa = 9.96KK571 pKa = 10.75GGGQDD576 pKa = 3.01EE577 pKa = 4.91SEE579 pKa = 4.29SHH581 pKa = 6.53RR582 pKa = 11.84EE583 pKa = 3.46ISGRR587 pKa = 11.84DD588 pKa = 3.38PGRR591 pKa = 11.84EE592 pKa = 4.14SNDD595 pKa = 3.04KK596 pKa = 10.79SPQGEE601 pKa = 3.89AEE603 pKa = 3.75EE604 pKa = 4.24FEE606 pKa = 5.37RR607 pKa = 11.84YY608 pKa = 9.07FSSFYY613 pKa = 10.49SWKK616 pKa = 9.96LHH618 pKa = 6.1NSGEE622 pKa = 4.38ANSGFRR628 pKa = 11.84PCGKK632 pKa = 9.13IPKK635 pKa = 9.66FYY637 pKa = 10.38RR638 pKa = 11.84PRR640 pKa = 11.84KK641 pKa = 8.78RR642 pKa = 11.84RR643 pKa = 11.84VSEE646 pKa = 3.54WGQNLARR653 pKa = 11.84KK654 pKa = 9.15HH655 pKa = 6.01SSLGEE660 pKa = 3.73ITQGFGWPEE669 pKa = 3.48AGAEE673 pKa = 4.04AEE675 pKa = 4.15LRR677 pKa = 11.84SLRR680 pKa = 11.84LQAQRR685 pKa = 11.84WLEE688 pKa = 3.84RR689 pKa = 11.84SKK691 pKa = 11.51SSVIPSAIEE700 pKa = 3.64RR701 pKa = 11.84EE702 pKa = 4.27IVISRR707 pKa = 11.84LVEE710 pKa = 3.94SYY712 pKa = 10.48KK713 pKa = 9.93ICRR716 pKa = 11.84SEE718 pKa = 5.19APLCSSGSDD727 pKa = 3.62LSWKK731 pKa = 10.44GFLEE735 pKa = 4.19DD736 pKa = 3.46FRR738 pKa = 11.84EE739 pKa = 4.44AVSSLEE745 pKa = 3.78LDD747 pKa = 3.39AGIGVPYY754 pKa = 9.94IGYY757 pKa = 9.7GYY759 pKa = 6.91PTHH762 pKa = 7.0RR763 pKa = 11.84GWVEE767 pKa = 3.72NPRR770 pKa = 11.84LLPVLSRR777 pKa = 11.84LVYY780 pKa = 10.69ARR782 pKa = 11.84LQRR785 pKa = 11.84LATLSVDD792 pKa = 3.11GKK794 pKa = 9.08TPEE797 pKa = 3.92EE798 pKa = 4.07LVRR801 pKa = 11.84DD802 pKa = 3.98GLVDD806 pKa = 3.64PVRR809 pKa = 11.84VFVKK813 pKa = 10.77GEE815 pKa = 3.59PHH817 pKa = 6.19KK818 pKa = 10.54QSKK821 pKa = 10.21LDD823 pKa = 3.38EE824 pKa = 4.27GRR826 pKa = 11.84YY827 pKa = 8.96RR828 pKa = 11.84LIMSVSLIDD837 pKa = 3.51QLVARR842 pKa = 11.84VLFQKK847 pKa = 10.13QNKK850 pKa = 9.68LEE852 pKa = 4.19LLLWRR857 pKa = 11.84SIPSKK862 pKa = 10.52PGFGLSTVEE871 pKa = 3.97QVEE874 pKa = 4.17EE875 pKa = 4.9FIDD878 pKa = 3.42HH879 pKa = 7.41LARR882 pKa = 11.84VVDD885 pKa = 4.2VKK887 pKa = 11.37SDD889 pKa = 3.68DD890 pKa = 4.52LLEE893 pKa = 4.12NWRR896 pKa = 11.84EE897 pKa = 3.86LMVPTDD903 pKa = 3.69CSGFDD908 pKa = 3.34WSVSDD913 pKa = 4.32WMLKK917 pKa = 10.91DD918 pKa = 3.36EE919 pKa = 4.34MEE921 pKa = 4.09VRR923 pKa = 11.84NRR925 pKa = 11.84LTINCNDD932 pKa = 3.23LTRR935 pKa = 11.84RR936 pKa = 11.84LRR938 pKa = 11.84NSWLYY943 pKa = 10.75CLSNSDD949 pKa = 4.24LALSDD954 pKa = 3.77GSLLAQEE961 pKa = 4.54VPGVQKK967 pKa = 10.68SGSYY971 pKa = 8.27NTSSTNSRR979 pKa = 11.84IRR981 pKa = 11.84VMAAYY986 pKa = 9.45FAGASWAVAIGDD998 pKa = 4.3DD999 pKa = 3.91ALEE1002 pKa = 4.98SIDD1005 pKa = 3.67TTLAVYY1011 pKa = 10.38KK1012 pKa = 10.72SLGFKK1017 pKa = 10.76VEE1019 pKa = 3.91VSEE1022 pKa = 5.07DD1023 pKa = 3.72LEE1025 pKa = 4.48FCSHH1029 pKa = 6.22IFKK1032 pKa = 9.77TRR1034 pKa = 11.84SLAIPVNTSKK1044 pKa = 10.24MLYY1047 pKa = 9.31RR1048 pKa = 11.84LIYY1051 pKa = 10.06GYY1053 pKa = 10.57EE1054 pKa = 4.09PEE1056 pKa = 5.02CGNLDD1061 pKa = 3.42VLRR1064 pKa = 11.84NYY1066 pKa = 10.64LCALASVLHH1075 pKa = 6.07EE1076 pKa = 4.51LRR1078 pKa = 11.84HH1079 pKa = 6.23DD1080 pKa = 3.55QDD1082 pKa = 4.09LVQNLSKK1089 pKa = 10.44WLIPDD1094 pKa = 4.33GSQKK1098 pKa = 10.24ISS1100 pKa = 3.08
Molecular weight: 122.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q5NDM9|RDRP_PNLV Protein P2-P3 OS=Poinsettia latent virus (isolate Euphorbia pulcherrima/Germany/Siepen/2005) OX=686943 GN=ORF2/ORF3 PE=3 SV=2
MM1 pKa = 7.52AKK3 pKa = 10.28KK4 pKa = 10.22GVNKK8 pKa = 9.38QARR11 pKa = 11.84TNSKK15 pKa = 10.23LDD17 pKa = 3.36QLVTSLEE24 pKa = 4.07RR25 pKa = 11.84LTVGSMQRR33 pKa = 11.84KK34 pKa = 8.92SNRR37 pKa = 11.84QRR39 pKa = 11.84PKK41 pKa = 9.87SRR43 pKa = 11.84KK44 pKa = 6.78TSGRR48 pKa = 11.84VVTAPVSSAALIRR61 pKa = 11.84YY62 pKa = 7.55KK63 pKa = 10.82APSLNGNSMDD73 pKa = 4.08SVRR76 pKa = 11.84LASKK80 pKa = 10.47SLVIEE85 pKa = 3.96AVKK88 pKa = 10.84VGIEE92 pKa = 4.08YY93 pKa = 10.25QVSVVPLVPCIQHH106 pKa = 5.29TWLSGVACYY115 pKa = 8.8WSKK118 pKa = 11.04YY119 pKa = 6.48RR120 pKa = 11.84WRR122 pKa = 11.84SCRR125 pKa = 11.84LVYY128 pKa = 10.09IPYY131 pKa = 9.67CSSSFTGYY139 pKa = 10.88VSMGLSYY146 pKa = 10.82DD147 pKa = 3.66YY148 pKa = 11.71ADD150 pKa = 5.83AIPSDD155 pKa = 4.08EE156 pKa = 4.53TEE158 pKa = 3.76MSALKK163 pKa = 10.69GYY165 pKa = 6.35TTTSAWAGSEE175 pKa = 4.22GIKK178 pKa = 9.62MLSYY182 pKa = 8.7PTGAVPSGAVVLSLDD197 pKa = 4.2CASMSKK203 pKa = 9.83PWYY206 pKa = 8.78PYY208 pKa = 9.39ISAVAATNLGEE219 pKa = 4.28KK220 pKa = 10.23QPEE223 pKa = 4.43LLNQYY228 pKa = 8.72TPARR232 pKa = 11.84AFIGTGGGMQGVNGLTAGRR251 pKa = 11.84VFMVYY256 pKa = 9.97DD257 pKa = 3.63IEE259 pKa = 5.48LIEE262 pKa = 4.27PVSPAMQEE270 pKa = 4.16NLSSTGRR277 pKa = 11.84SKK279 pKa = 10.44IDD281 pKa = 3.29QSISPLTGADD291 pKa = 3.46FEE293 pKa = 4.62IVRR296 pKa = 11.84KK297 pKa = 10.07VSAISQIQQ305 pKa = 2.93
MM1 pKa = 7.52AKK3 pKa = 10.28KK4 pKa = 10.22GVNKK8 pKa = 9.38QARR11 pKa = 11.84TNSKK15 pKa = 10.23LDD17 pKa = 3.36QLVTSLEE24 pKa = 4.07RR25 pKa = 11.84LTVGSMQRR33 pKa = 11.84KK34 pKa = 8.92SNRR37 pKa = 11.84QRR39 pKa = 11.84PKK41 pKa = 9.87SRR43 pKa = 11.84KK44 pKa = 6.78TSGRR48 pKa = 11.84VVTAPVSSAALIRR61 pKa = 11.84YY62 pKa = 7.55KK63 pKa = 10.82APSLNGNSMDD73 pKa = 4.08SVRR76 pKa = 11.84LASKK80 pKa = 10.47SLVIEE85 pKa = 3.96AVKK88 pKa = 10.84VGIEE92 pKa = 4.08YY93 pKa = 10.25QVSVVPLVPCIQHH106 pKa = 5.29TWLSGVACYY115 pKa = 8.8WSKK118 pKa = 11.04YY119 pKa = 6.48RR120 pKa = 11.84WRR122 pKa = 11.84SCRR125 pKa = 11.84LVYY128 pKa = 10.09IPYY131 pKa = 9.67CSSSFTGYY139 pKa = 10.88VSMGLSYY146 pKa = 10.82DD147 pKa = 3.66YY148 pKa = 11.71ADD150 pKa = 5.83AIPSDD155 pKa = 4.08EE156 pKa = 4.53TEE158 pKa = 3.76MSALKK163 pKa = 10.69GYY165 pKa = 6.35TTTSAWAGSEE175 pKa = 4.22GIKK178 pKa = 9.62MLSYY182 pKa = 8.7PTGAVPSGAVVLSLDD197 pKa = 4.2CASMSKK203 pKa = 9.83PWYY206 pKa = 8.78PYY208 pKa = 9.39ISAVAATNLGEE219 pKa = 4.28KK220 pKa = 10.23QPEE223 pKa = 4.43LLNQYY228 pKa = 8.72TPARR232 pKa = 11.84AFIGTGGGMQGVNGLTAGRR251 pKa = 11.84VFMVYY256 pKa = 9.97DD257 pKa = 3.63IEE259 pKa = 5.48LIEE262 pKa = 4.27PVSPAMQEE270 pKa = 4.16NLSSTGRR277 pKa = 11.84SKK279 pKa = 10.44IDD281 pKa = 3.29QSISPLTGADD291 pKa = 3.46FEE293 pKa = 4.62IVRR296 pKa = 11.84KK297 pKa = 10.07VSAISQIQQ305 pKa = 2.93
Molecular weight: 33.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2331 |
266 |
1100 |
582.8 |
64.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.049 ± 0.572 | 2.66 ± 0.348 |
4.462 ± 0.335 | 5.577 ± 0.776 |
3.175 ± 0.538 | 7.293 ± 0.429 |
1.931 ± 0.32 | 5.448 ± 0.284 |
6.006 ± 0.707 | 10.167 ± 0.609 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.231 ± 0.233 | 4.161 ± 0.234 |
5.32 ± 0.316 | 2.574 ± 0.426 |
5.32 ± 0.558 | 10.854 ± 0.39 |
4.419 ± 0.505 | 6.821 ± 0.477 |
2.188 ± 0.124 | 3.346 ± 0.438 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
