
Candidatus Magnetoglobus multicellularis str. Araruama
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae; Candidatus Magnetoglobus; Candidatus Magnetoglobus multicellularis
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
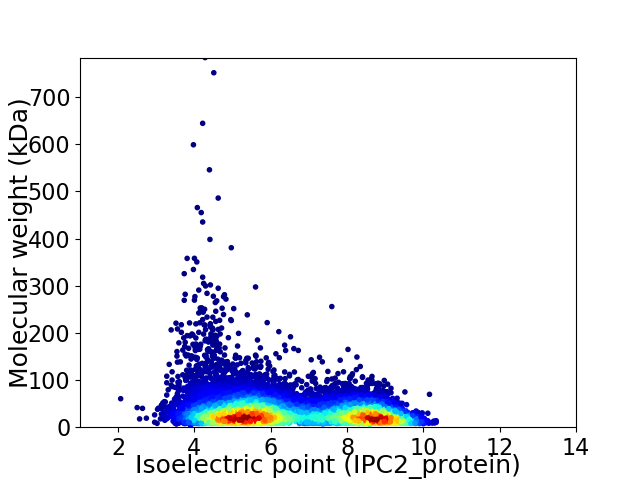
Virtual 2D-PAGE plot for 9976 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
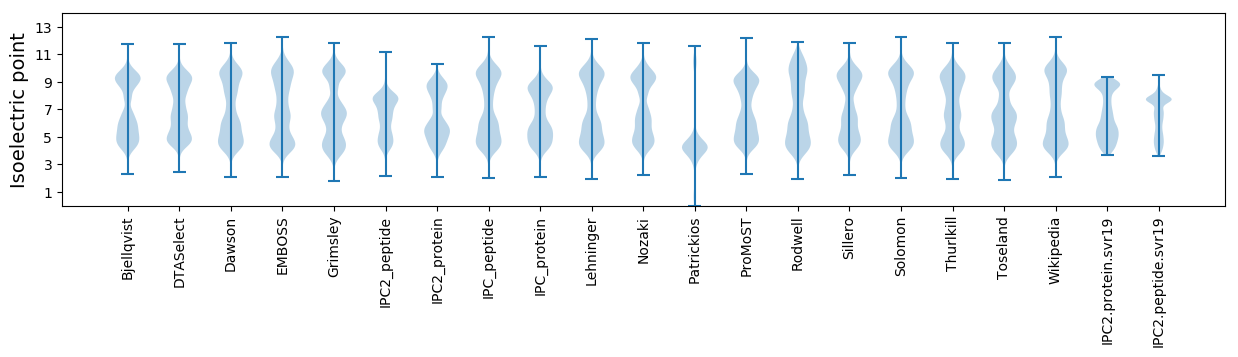
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1V1NSZ5|A0A1V1NSZ5_9DELT Glutamine--scyllo-inositol transaminase (Fragment) OS=Candidatus Magnetoglobus multicellularis str. Araruama OX=890399 GN=OMM_13869 PE=4 SV=1
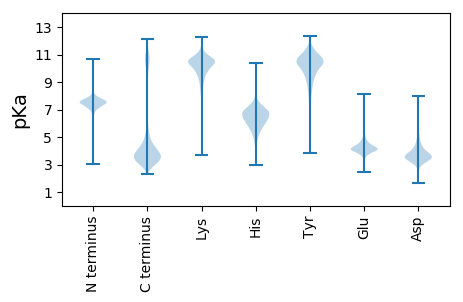
AA1 pKa = 7.55FADD4 pKa = 3.82YY5 pKa = 11.36DD6 pKa = 3.86NDD8 pKa = 3.76GDD10 pKa = 4.82LDD12 pKa = 3.74ILITGDD18 pKa = 3.38SDD20 pKa = 3.33NGKK23 pKa = 8.01IAKK26 pKa = 8.11MFRR29 pKa = 11.84NTGNAFTEE37 pKa = 4.1DD38 pKa = 2.96TGINLTGVSHH48 pKa = 7.13SSTAFGDD55 pKa = 3.58YY56 pKa = 11.14DD57 pKa = 4.16NDD59 pKa = 3.76GDD61 pKa = 5.21LDD63 pKa = 4.15LLITGRR69 pKa = 11.84SDD71 pKa = 3.34SLIIAKK77 pKa = 9.17MYY79 pKa = 10.75QNTGGNFSEE88 pKa = 4.55DD89 pKa = 3.16TTINLMGVEE98 pKa = 4.26YY99 pKa = 10.65GSTSFGDD106 pKa = 3.56YY107 pKa = 11.22DD108 pKa = 4.13NDD110 pKa = 3.69GDD112 pKa = 5.0LDD114 pKa = 3.82ILISGDD120 pKa = 3.15TGSNKK125 pKa = 7.64ITKK128 pKa = 9.65VYY130 pKa = 10.36RR131 pKa = 11.84NTGGSFSEE139 pKa = 4.27DD140 pKa = 2.97TGIHH144 pKa = 6.76LIGVFDD150 pKa = 4.03GASAFGDD157 pKa = 3.7YY158 pKa = 11.26DD159 pKa = 3.87NDD161 pKa = 3.71GDD163 pKa = 5.07LDD165 pKa = 3.76ILITGDD171 pKa = 3.21TGNSKK176 pKa = 8.52NTTVYY181 pKa = 10.9QNTDD185 pKa = 2.67GHH187 pKa = 6.37FSEE190 pKa = 6.03ASDD193 pKa = 4.03INLTDD198 pKa = 3.55VTHH201 pKa = 5.83STTVFGDD208 pKa = 3.33YY209 pKa = 11.18DD210 pKa = 4.0NDD212 pKa = 3.83GDD214 pKa = 5.59LDD216 pKa = 3.75ILMSGQAASGRR227 pKa = 11.84MAKK230 pKa = 9.8IYY232 pKa = 10.51QNNTLISNTPANAPSSLTSVVSEE255 pKa = 4.2QIVLLSWSAASDD267 pKa = 4.01SEE269 pKa = 4.45TLSAAGLSYY278 pKa = 11.08NLRR281 pKa = 11.84IGSSPGACDD290 pKa = 3.09ILSPMALPLSNGYY303 pKa = 8.95RR304 pKa = 11.84QIPARR309 pKa = 11.84GAIQTLTATINPLNGGTYY327 pKa = 7.97YY328 pKa = 10.33WSVQSIDD335 pKa = 3.39TAFAGSDD342 pKa = 3.41FSNEE346 pKa = 3.77STFTIDD352 pKa = 2.97QTPIISAIAHH362 pKa = 4.54QHH364 pKa = 4.41TVIDD368 pKa = 4.14YY369 pKa = 7.81PISIPFQLL377 pKa = 4.32
AA1 pKa = 7.55FADD4 pKa = 3.82YY5 pKa = 11.36DD6 pKa = 3.86NDD8 pKa = 3.76GDD10 pKa = 4.82LDD12 pKa = 3.74ILITGDD18 pKa = 3.38SDD20 pKa = 3.33NGKK23 pKa = 8.01IAKK26 pKa = 8.11MFRR29 pKa = 11.84NTGNAFTEE37 pKa = 4.1DD38 pKa = 2.96TGINLTGVSHH48 pKa = 7.13SSTAFGDD55 pKa = 3.58YY56 pKa = 11.14DD57 pKa = 4.16NDD59 pKa = 3.76GDD61 pKa = 5.21LDD63 pKa = 4.15LLITGRR69 pKa = 11.84SDD71 pKa = 3.34SLIIAKK77 pKa = 9.17MYY79 pKa = 10.75QNTGGNFSEE88 pKa = 4.55DD89 pKa = 3.16TTINLMGVEE98 pKa = 4.26YY99 pKa = 10.65GSTSFGDD106 pKa = 3.56YY107 pKa = 11.22DD108 pKa = 4.13NDD110 pKa = 3.69GDD112 pKa = 5.0LDD114 pKa = 3.82ILISGDD120 pKa = 3.15TGSNKK125 pKa = 7.64ITKK128 pKa = 9.65VYY130 pKa = 10.36RR131 pKa = 11.84NTGGSFSEE139 pKa = 4.27DD140 pKa = 2.97TGIHH144 pKa = 6.76LIGVFDD150 pKa = 4.03GASAFGDD157 pKa = 3.7YY158 pKa = 11.26DD159 pKa = 3.87NDD161 pKa = 3.71GDD163 pKa = 5.07LDD165 pKa = 3.76ILITGDD171 pKa = 3.21TGNSKK176 pKa = 8.52NTTVYY181 pKa = 10.9QNTDD185 pKa = 2.67GHH187 pKa = 6.37FSEE190 pKa = 6.03ASDD193 pKa = 4.03INLTDD198 pKa = 3.55VTHH201 pKa = 5.83STTVFGDD208 pKa = 3.33YY209 pKa = 11.18DD210 pKa = 4.0NDD212 pKa = 3.83GDD214 pKa = 5.59LDD216 pKa = 3.75ILMSGQAASGRR227 pKa = 11.84MAKK230 pKa = 9.8IYY232 pKa = 10.51QNNTLISNTPANAPSSLTSVVSEE255 pKa = 4.2QIVLLSWSAASDD267 pKa = 4.01SEE269 pKa = 4.45TLSAAGLSYY278 pKa = 11.08NLRR281 pKa = 11.84IGSSPGACDD290 pKa = 3.09ILSPMALPLSNGYY303 pKa = 8.95RR304 pKa = 11.84QIPARR309 pKa = 11.84GAIQTLTATINPLNGGTYY327 pKa = 7.97YY328 pKa = 10.33WSVQSIDD335 pKa = 3.39TAFAGSDD342 pKa = 3.41FSNEE346 pKa = 3.77STFTIDD352 pKa = 2.97QTPIISAIAHH362 pKa = 4.54QHH364 pKa = 4.41TVIDD368 pKa = 4.14YY369 pKa = 7.81PISIPFQLL377 pKa = 4.32
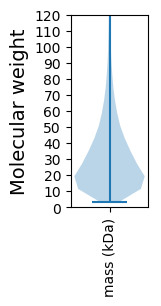
Molecular weight: 39.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1V1NVQ3|A0A1V1NVQ3_9DELT Uncharacterized protein (Fragment) OS=Candidatus Magnetoglobus multicellularis str. Araruama OX=890399 GN=OMM_12535 PE=4 SV=1
MM1 pKa = 7.81ASFEE5 pKa = 4.65KK6 pKa = 10.49PLEE9 pKa = 4.19TRR11 pKa = 11.84FLRR14 pKa = 11.84PSQRR18 pKa = 11.84KK19 pKa = 7.17LTASFTLSKK28 pKa = 10.41CLYY31 pKa = 8.92SWYY34 pKa = 10.63SPAARR39 pKa = 11.84AATPMIVAPTGLAARR54 pKa = 11.84VMRR57 pKa = 11.84DD58 pKa = 3.35VPTMPIAPLSMPRR71 pKa = 11.84PDD73 pKa = 3.69VSAANNGPAVARR85 pKa = 11.84PAAITPYY92 pKa = 9.88AAPAAPITATTVPAAAATPVSPIAIFVSMGCAVMKK127 pKa = 9.81PATLFAIFKK136 pKa = 10.19IIVPTGARR144 pKa = 11.84ATATLAADD152 pKa = 4.61LMNSLLTVRR161 pKa = 11.84HH162 pKa = 6.33ASPSSRR168 pKa = 11.84VRR170 pKa = 11.84VLDD173 pKa = 3.56VSNCRR178 pKa = 11.84FNVPEE183 pKa = 4.46TFPSTLSGDD192 pKa = 3.46
MM1 pKa = 7.81ASFEE5 pKa = 4.65KK6 pKa = 10.49PLEE9 pKa = 4.19TRR11 pKa = 11.84FLRR14 pKa = 11.84PSQRR18 pKa = 11.84KK19 pKa = 7.17LTASFTLSKK28 pKa = 10.41CLYY31 pKa = 8.92SWYY34 pKa = 10.63SPAARR39 pKa = 11.84AATPMIVAPTGLAARR54 pKa = 11.84VMRR57 pKa = 11.84DD58 pKa = 3.35VPTMPIAPLSMPRR71 pKa = 11.84PDD73 pKa = 3.69VSAANNGPAVARR85 pKa = 11.84PAAITPYY92 pKa = 9.88AAPAAPITATTVPAAAATPVSPIAIFVSMGCAVMKK127 pKa = 9.81PATLFAIFKK136 pKa = 10.19IIVPTGARR144 pKa = 11.84ATATLAADD152 pKa = 4.61LMNSLLTVRR161 pKa = 11.84HH162 pKa = 6.33ASPSSRR168 pKa = 11.84VRR170 pKa = 11.84VLDD173 pKa = 3.56VSNCRR178 pKa = 11.84FNVPEE183 pKa = 4.46TFPSTLSGDD192 pKa = 3.46
Molecular weight: 20.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3167227 |
32 |
7151 |
317.5 |
35.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.941 ± 0.027 | 1.199 ± 0.009 |
6.279 ± 0.02 | 5.698 ± 0.024 |
4.495 ± 0.019 | 5.95 ± 0.029 |
2.249 ± 0.012 | 8.781 ± 0.022 |
6.399 ± 0.04 | 8.762 ± 0.032 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.445 ± 0.015 | 5.95 ± 0.023 |
3.825 ± 0.018 | 4.303 ± 0.018 |
3.783 ± 0.024 | 7.336 ± 0.037 |
5.983 ± 0.043 | 5.568 ± 0.021 |
1.164 ± 0.01 | 3.888 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
