
Caldithrix abyssi DSM 13497
Taxonomy: cellular organisms; Bacteria; Calditrichaeota; Calditrichia; Calditrichales; Calditrichaceae; Caldithrix; Caldithrix abyssi
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
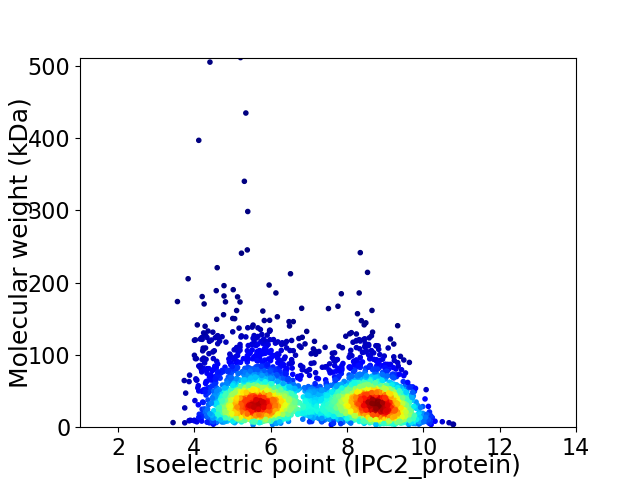
Virtual 2D-PAGE plot for 3674 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H1XP64|H1XP64_9BACT Amino acid adenylation domain protein OS=Caldithrix abyssi DSM 13497 OX=880073 GN=Cabys_1398 PE=4 SV=1
MM1 pKa = 7.14KK2 pKa = 9.69TRR4 pKa = 11.84IVILILGLATAILATTYY21 pKa = 8.31HH22 pKa = 6.33TLPFNGQNAFAPDD35 pKa = 3.55EE36 pKa = 4.37DD37 pKa = 5.7FITSTDD43 pKa = 3.3TVLAYY48 pKa = 7.5CTWDD52 pKa = 3.35DD53 pKa = 3.41QYY55 pKa = 11.93LYY57 pKa = 10.74LGLSSPFLATPNDD70 pKa = 4.05TARR73 pKa = 11.84GRR75 pKa = 11.84YY76 pKa = 9.58DD77 pKa = 3.49SFWYY81 pKa = 9.87IDD83 pKa = 3.96TDD85 pKa = 3.59PHH87 pKa = 6.64PDD89 pKa = 3.38NPKK92 pKa = 10.53SGLGTDD98 pKa = 3.22QTGSYY103 pKa = 9.68FIQINTSQPWWFDD116 pKa = 3.42EE117 pKa = 4.41QSWEE121 pKa = 4.11LPFFADD127 pKa = 3.23YY128 pKa = 10.84RR129 pKa = 11.84IIPRR133 pKa = 11.84YY134 pKa = 7.97YY135 pKa = 10.72KK136 pKa = 10.08KK137 pKa = 10.75DD138 pKa = 3.14SVYY141 pKa = 11.3ASFYY145 pKa = 11.06YY146 pKa = 10.4YY147 pKa = 10.82DD148 pKa = 3.83SDD150 pKa = 3.68SARR153 pKa = 11.84WFFRR157 pKa = 11.84GDD159 pKa = 2.96IDD161 pKa = 4.45TSLANLNYY169 pKa = 9.37IDD171 pKa = 4.74GYY173 pKa = 10.74YY174 pKa = 10.5EE175 pKa = 4.07LRR177 pKa = 11.84LPLDD181 pKa = 3.62SLNFPTDD188 pKa = 3.32INILGYY194 pKa = 10.39SVDD197 pKa = 4.21GQWEE201 pKa = 4.15SDD203 pKa = 3.64IYY205 pKa = 10.84WDD207 pKa = 4.72DD208 pKa = 3.64EE209 pKa = 4.67YY210 pKa = 11.73GFTRR214 pKa = 11.84DD215 pKa = 3.09VGGTFASWPWSSLVGGDD232 pKa = 3.98GDD234 pKa = 4.06YY235 pKa = 9.27HH236 pKa = 5.9TKK238 pKa = 10.8GKK240 pKa = 9.79FNHH243 pKa = 6.72WFHH246 pKa = 6.76FHH248 pKa = 6.54LQPGISPDD256 pKa = 3.64QEE258 pKa = 3.82NDD260 pKa = 3.45PPVASPITGQTINEE274 pKa = 4.32GEE276 pKa = 4.33SFAVIDD282 pKa = 4.05LNSYY286 pKa = 10.37VFDD289 pKa = 5.45DD290 pKa = 4.5LTPDD294 pKa = 3.45TLLIWTTEE302 pKa = 4.2TIDD305 pKa = 5.48LIVTIQDD312 pKa = 3.35SNQAQVSTPNPNWNGTEE329 pKa = 4.08TITFIVTDD337 pKa = 4.04EE338 pKa = 4.51GGKK341 pKa = 10.03SDD343 pKa = 3.69TTTGTFTMLGDD354 pKa = 3.61NQIPVAVNDD363 pKa = 3.6SSNTIEE369 pKa = 4.19EE370 pKa = 4.04QSVSINLTANDD381 pKa = 3.81SDD383 pKa = 4.31ADD385 pKa = 3.92NDD387 pKa = 4.01SLHH390 pKa = 7.2IDD392 pKa = 4.85RR393 pKa = 11.84ILTTNNGTVVIDD405 pKa = 3.62NDD407 pKa = 3.85SMVTYY412 pKa = 9.28TPNVDD417 pKa = 4.01FHH419 pKa = 6.39GTDD422 pKa = 2.86SFEE425 pKa = 4.39YY426 pKa = 10.85VVTDD430 pKa = 3.68GNGGRR435 pKa = 11.84DD436 pKa = 3.23TATVFVTVDD445 pKa = 3.25NRR447 pKa = 11.84NDD449 pKa = 3.32PPEE452 pKa = 4.39IVNLPDD458 pKa = 4.06VINMTTNDD466 pKa = 3.38STKK469 pKa = 10.71LYY471 pKa = 9.72MMDD474 pKa = 3.13YY475 pKa = 10.21AYY477 pKa = 10.61DD478 pKa = 3.54VDD480 pKa = 4.52TPDD483 pKa = 5.8SLLTWSFSVNDD494 pKa = 3.87PAISYY499 pKa = 10.45AYY501 pKa = 10.25DD502 pKa = 3.24QTTDD506 pKa = 2.78TLTIYY511 pKa = 10.41SHH513 pKa = 7.2EE514 pKa = 4.48INGDD518 pKa = 3.65FYY520 pKa = 11.69LFTTLTDD527 pKa = 3.75DD528 pKa = 5.31SGASDD533 pKa = 3.77QDD535 pKa = 3.95TITIRR540 pKa = 11.84VSGTSALDD548 pKa = 3.6LRR550 pKa = 11.84STAIPDD556 pKa = 3.42QFTVLQNFPNPFNPSTHH573 pKa = 6.47LAFGLTKK580 pKa = 10.65SSDD583 pKa = 3.48VQIEE587 pKa = 3.94IFNILGQRR595 pKa = 11.84VFSKK599 pKa = 10.59RR600 pKa = 11.84LSGLSAGFHH609 pKa = 4.98TVKK612 pKa = 10.24VDD614 pKa = 3.41AARR617 pKa = 11.84WPAGIYY623 pKa = 10.06LYY625 pKa = 10.41RR626 pKa = 11.84ISAEE630 pKa = 3.55NTMVIKK636 pKa = 10.85KK637 pKa = 8.83MVLVRR642 pKa = 4.15
MM1 pKa = 7.14KK2 pKa = 9.69TRR4 pKa = 11.84IVILILGLATAILATTYY21 pKa = 8.31HH22 pKa = 6.33TLPFNGQNAFAPDD35 pKa = 3.55EE36 pKa = 4.37DD37 pKa = 5.7FITSTDD43 pKa = 3.3TVLAYY48 pKa = 7.5CTWDD52 pKa = 3.35DD53 pKa = 3.41QYY55 pKa = 11.93LYY57 pKa = 10.74LGLSSPFLATPNDD70 pKa = 4.05TARR73 pKa = 11.84GRR75 pKa = 11.84YY76 pKa = 9.58DD77 pKa = 3.49SFWYY81 pKa = 9.87IDD83 pKa = 3.96TDD85 pKa = 3.59PHH87 pKa = 6.64PDD89 pKa = 3.38NPKK92 pKa = 10.53SGLGTDD98 pKa = 3.22QTGSYY103 pKa = 9.68FIQINTSQPWWFDD116 pKa = 3.42EE117 pKa = 4.41QSWEE121 pKa = 4.11LPFFADD127 pKa = 3.23YY128 pKa = 10.84RR129 pKa = 11.84IIPRR133 pKa = 11.84YY134 pKa = 7.97YY135 pKa = 10.72KK136 pKa = 10.08KK137 pKa = 10.75DD138 pKa = 3.14SVYY141 pKa = 11.3ASFYY145 pKa = 11.06YY146 pKa = 10.4YY147 pKa = 10.82DD148 pKa = 3.83SDD150 pKa = 3.68SARR153 pKa = 11.84WFFRR157 pKa = 11.84GDD159 pKa = 2.96IDD161 pKa = 4.45TSLANLNYY169 pKa = 9.37IDD171 pKa = 4.74GYY173 pKa = 10.74YY174 pKa = 10.5EE175 pKa = 4.07LRR177 pKa = 11.84LPLDD181 pKa = 3.62SLNFPTDD188 pKa = 3.32INILGYY194 pKa = 10.39SVDD197 pKa = 4.21GQWEE201 pKa = 4.15SDD203 pKa = 3.64IYY205 pKa = 10.84WDD207 pKa = 4.72DD208 pKa = 3.64EE209 pKa = 4.67YY210 pKa = 11.73GFTRR214 pKa = 11.84DD215 pKa = 3.09VGGTFASWPWSSLVGGDD232 pKa = 3.98GDD234 pKa = 4.06YY235 pKa = 9.27HH236 pKa = 5.9TKK238 pKa = 10.8GKK240 pKa = 9.79FNHH243 pKa = 6.72WFHH246 pKa = 6.76FHH248 pKa = 6.54LQPGISPDD256 pKa = 3.64QEE258 pKa = 3.82NDD260 pKa = 3.45PPVASPITGQTINEE274 pKa = 4.32GEE276 pKa = 4.33SFAVIDD282 pKa = 4.05LNSYY286 pKa = 10.37VFDD289 pKa = 5.45DD290 pKa = 4.5LTPDD294 pKa = 3.45TLLIWTTEE302 pKa = 4.2TIDD305 pKa = 5.48LIVTIQDD312 pKa = 3.35SNQAQVSTPNPNWNGTEE329 pKa = 4.08TITFIVTDD337 pKa = 4.04EE338 pKa = 4.51GGKK341 pKa = 10.03SDD343 pKa = 3.69TTTGTFTMLGDD354 pKa = 3.61NQIPVAVNDD363 pKa = 3.6SSNTIEE369 pKa = 4.19EE370 pKa = 4.04QSVSINLTANDD381 pKa = 3.81SDD383 pKa = 4.31ADD385 pKa = 3.92NDD387 pKa = 4.01SLHH390 pKa = 7.2IDD392 pKa = 4.85RR393 pKa = 11.84ILTTNNGTVVIDD405 pKa = 3.62NDD407 pKa = 3.85SMVTYY412 pKa = 9.28TPNVDD417 pKa = 4.01FHH419 pKa = 6.39GTDD422 pKa = 2.86SFEE425 pKa = 4.39YY426 pKa = 10.85VVTDD430 pKa = 3.68GNGGRR435 pKa = 11.84DD436 pKa = 3.23TATVFVTVDD445 pKa = 3.25NRR447 pKa = 11.84NDD449 pKa = 3.32PPEE452 pKa = 4.39IVNLPDD458 pKa = 4.06VINMTTNDD466 pKa = 3.38STKK469 pKa = 10.71LYY471 pKa = 9.72MMDD474 pKa = 3.13YY475 pKa = 10.21AYY477 pKa = 10.61DD478 pKa = 3.54VDD480 pKa = 4.52TPDD483 pKa = 5.8SLLTWSFSVNDD494 pKa = 3.87PAISYY499 pKa = 10.45AYY501 pKa = 10.25DD502 pKa = 3.24QTTDD506 pKa = 2.78TLTIYY511 pKa = 10.41SHH513 pKa = 7.2EE514 pKa = 4.48INGDD518 pKa = 3.65FYY520 pKa = 11.69LFTTLTDD527 pKa = 3.75DD528 pKa = 5.31SGASDD533 pKa = 3.77QDD535 pKa = 3.95TITIRR540 pKa = 11.84VSGTSALDD548 pKa = 3.6LRR550 pKa = 11.84STAIPDD556 pKa = 3.42QFTVLQNFPNPFNPSTHH573 pKa = 6.47LAFGLTKK580 pKa = 10.65SSDD583 pKa = 3.48VQIEE587 pKa = 3.94IFNILGQRR595 pKa = 11.84VFSKK599 pKa = 10.59RR600 pKa = 11.84LSGLSAGFHH609 pKa = 4.98TVKK612 pKa = 10.24VDD614 pKa = 3.41AARR617 pKa = 11.84WPAGIYY623 pKa = 10.06LYY625 pKa = 10.41RR626 pKa = 11.84ISAEE630 pKa = 3.55NTMVIKK636 pKa = 10.85KK637 pKa = 8.83MVLVRR642 pKa = 4.15
Molecular weight: 72.01 kDa
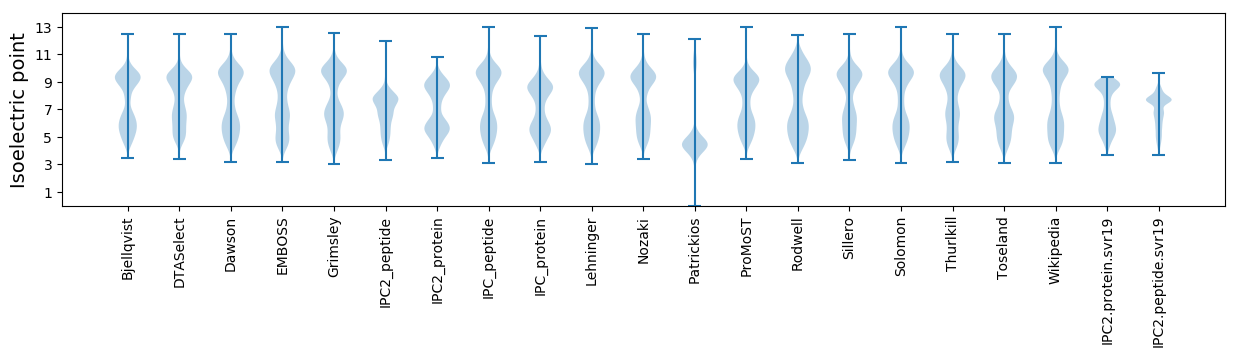
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H1XPH5|H1XPH5_9BACT Uncharacterized protein OS=Caldithrix abyssi DSM 13497 OX=880073 GN=Cabys_2709 PE=4 SV=1
MM1 pKa = 7.32PCGRR5 pKa = 11.84KK6 pKa = 9.29RR7 pKa = 11.84KK8 pKa = 7.42RR9 pKa = 11.84AKK11 pKa = 9.56MATHH15 pKa = 6.33KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.16NRR25 pKa = 11.84HH26 pKa = 4.6KK27 pKa = 10.96KK28 pKa = 10.03KK29 pKa = 10.26IRR31 pKa = 3.43
MM1 pKa = 7.32PCGRR5 pKa = 11.84KK6 pKa = 9.29RR7 pKa = 11.84KK8 pKa = 7.42RR9 pKa = 11.84AKK11 pKa = 9.56MATHH15 pKa = 6.33KK16 pKa = 10.31RR17 pKa = 11.84KK18 pKa = 9.84KK19 pKa = 9.28RR20 pKa = 11.84LRR22 pKa = 11.84KK23 pKa = 9.16NRR25 pKa = 11.84HH26 pKa = 4.6KK27 pKa = 10.96KK28 pKa = 10.03KK29 pKa = 10.26IRR31 pKa = 3.43
Molecular weight: 3.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1397650 |
31 |
4710 |
380.4 |
43.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.103 ± 0.04 | 0.686 ± 0.013 |
5.294 ± 0.034 | 6.622 ± 0.044 |
5.251 ± 0.031 | 6.486 ± 0.038 |
2.016 ± 0.02 | 7.353 ± 0.035 |
6.686 ± 0.043 | 10.331 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.016 | 4.655 ± 0.032 |
4.136 ± 0.025 | 4.229 ± 0.029 |
5.096 ± 0.033 | 5.522 ± 0.031 |
4.81 ± 0.032 | 6.439 ± 0.031 |
1.249 ± 0.017 | 3.823 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
