
Wuhan arthropod virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
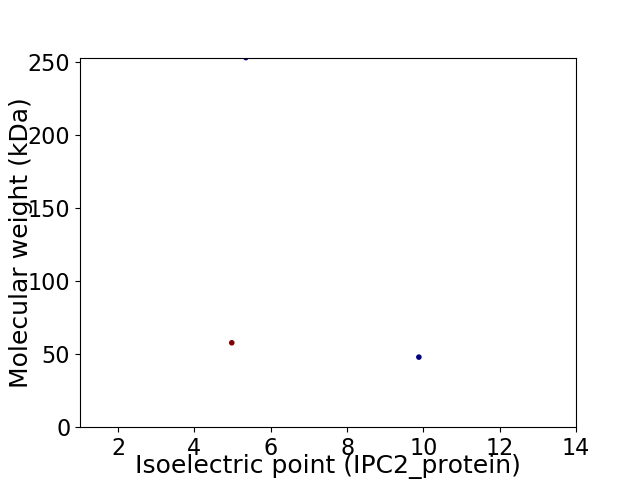
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KM75|A0A1L3KM75_9VIRU Calici_coat domain-containing protein OS=Wuhan arthropod virus 3 OX=1923692 PE=4 SV=1
MM1 pKa = 6.94STQTASAVSMGDD13 pKa = 3.23EE14 pKa = 4.32VFSSTEE20 pKa = 3.83DD21 pKa = 3.24APLPVSAIQAEE32 pKa = 4.39HH33 pKa = 7.24ADD35 pKa = 3.61PGEE38 pKa = 4.35AVPHH42 pKa = 5.7VGAANTIDD50 pKa = 3.46PFFYY54 pKa = 10.46EE55 pKa = 4.02QFVAQTNFTWTSSDD69 pKa = 3.47NPGKK73 pKa = 10.39LLYY76 pKa = 8.82TIPIHH81 pKa = 6.5PVFANIYY88 pKa = 9.38LAYY91 pKa = 10.36LVQLYY96 pKa = 7.27NTWVGGLDD104 pKa = 3.6FKK106 pKa = 11.66VKK108 pKa = 9.85VAGTGFHH115 pKa = 6.94AGALILARR123 pKa = 11.84IPPNIDD129 pKa = 3.2PNSLTTTSAITAFPCTFIDD148 pKa = 4.45PKK150 pKa = 10.31TLTAASQSIMDD161 pKa = 3.87QRR163 pKa = 11.84QLMYY167 pKa = 10.36HH168 pKa = 4.75YY169 pKa = 9.5TNYY172 pKa = 10.92DD173 pKa = 3.44SADD176 pKa = 3.32STTIGGHH183 pKa = 4.62FAIYY187 pKa = 10.2VLQSLNTSASGTNQIDD203 pKa = 3.58VQVFCKK209 pKa = 10.53ASADD213 pKa = 3.9FEE215 pKa = 4.52LLQIKK220 pKa = 9.32PPNLTLNPPTPTYY233 pKa = 11.69ANFTKK238 pKa = 10.75LFDD241 pKa = 3.96RR242 pKa = 11.84LAGQTTPYY250 pKa = 10.98AFGSNCIEE258 pKa = 4.8FIVAPTTGPIYY269 pKa = 10.69AGLEE273 pKa = 4.02GARR276 pKa = 11.84QLDD279 pKa = 3.93GSLLGTYY286 pKa = 7.36TTFDD290 pKa = 3.58EE291 pKa = 4.5QVTAQRR297 pKa = 11.84FSGISGVAGTNAGGKK312 pKa = 9.58LVIHH316 pKa = 6.93RR317 pKa = 11.84NAAAPYY323 pKa = 7.91TFQVIQFYY331 pKa = 11.04QNNCKK336 pKa = 8.5VTGVAMHH343 pKa = 7.38AIGDD347 pKa = 3.88EE348 pKa = 4.31STPASYY354 pKa = 10.94VSSVDD359 pKa = 3.11VDD361 pKa = 3.79FTLPYY366 pKa = 9.73NASSTAALGYY376 pKa = 9.19MDD378 pKa = 4.99EE379 pKa = 4.53SSATFVLTDD388 pKa = 4.49GINPVVIPTQGEE400 pKa = 4.42SLVGFGFYY408 pKa = 9.56ATRR411 pKa = 11.84SIAGSIYY418 pKa = 9.24YY419 pKa = 7.91TAQSTQMALGLRR431 pKa = 11.84DD432 pKa = 3.6KK433 pKa = 11.34VFGEE437 pKa = 4.38VPPDD441 pKa = 3.4SALLFILIDD450 pKa = 3.82TVLSIPVNYY459 pKa = 10.27LKK461 pKa = 10.73LYY463 pKa = 8.05PQGFFTSSHH472 pKa = 5.27VDD474 pKa = 3.16QPIVYY479 pKa = 9.97DD480 pKa = 3.36SRR482 pKa = 11.84RR483 pKa = 11.84YY484 pKa = 8.42RR485 pKa = 11.84LEE487 pKa = 3.64YY488 pKa = 10.35VGLIEE493 pKa = 4.41VTTTVPTKK501 pKa = 10.45IEE503 pKa = 4.18FEE505 pKa = 4.24THH507 pKa = 5.43KK508 pKa = 10.79LAMSRR513 pKa = 11.84SFDD516 pKa = 3.53HH517 pKa = 6.3KK518 pKa = 10.46KK519 pKa = 10.12KK520 pKa = 10.67RR521 pKa = 11.84SGFLSAAYY529 pKa = 9.71HH530 pKa = 6.2GEE532 pKa = 4.15SAAA535 pKa = 4.04
MM1 pKa = 6.94STQTASAVSMGDD13 pKa = 3.23EE14 pKa = 4.32VFSSTEE20 pKa = 3.83DD21 pKa = 3.24APLPVSAIQAEE32 pKa = 4.39HH33 pKa = 7.24ADD35 pKa = 3.61PGEE38 pKa = 4.35AVPHH42 pKa = 5.7VGAANTIDD50 pKa = 3.46PFFYY54 pKa = 10.46EE55 pKa = 4.02QFVAQTNFTWTSSDD69 pKa = 3.47NPGKK73 pKa = 10.39LLYY76 pKa = 8.82TIPIHH81 pKa = 6.5PVFANIYY88 pKa = 9.38LAYY91 pKa = 10.36LVQLYY96 pKa = 7.27NTWVGGLDD104 pKa = 3.6FKK106 pKa = 11.66VKK108 pKa = 9.85VAGTGFHH115 pKa = 6.94AGALILARR123 pKa = 11.84IPPNIDD129 pKa = 3.2PNSLTTTSAITAFPCTFIDD148 pKa = 4.45PKK150 pKa = 10.31TLTAASQSIMDD161 pKa = 3.87QRR163 pKa = 11.84QLMYY167 pKa = 10.36HH168 pKa = 4.75YY169 pKa = 9.5TNYY172 pKa = 10.92DD173 pKa = 3.44SADD176 pKa = 3.32STTIGGHH183 pKa = 4.62FAIYY187 pKa = 10.2VLQSLNTSASGTNQIDD203 pKa = 3.58VQVFCKK209 pKa = 10.53ASADD213 pKa = 3.9FEE215 pKa = 4.52LLQIKK220 pKa = 9.32PPNLTLNPPTPTYY233 pKa = 11.69ANFTKK238 pKa = 10.75LFDD241 pKa = 3.96RR242 pKa = 11.84LAGQTTPYY250 pKa = 10.98AFGSNCIEE258 pKa = 4.8FIVAPTTGPIYY269 pKa = 10.69AGLEE273 pKa = 4.02GARR276 pKa = 11.84QLDD279 pKa = 3.93GSLLGTYY286 pKa = 7.36TTFDD290 pKa = 3.58EE291 pKa = 4.5QVTAQRR297 pKa = 11.84FSGISGVAGTNAGGKK312 pKa = 9.58LVIHH316 pKa = 6.93RR317 pKa = 11.84NAAAPYY323 pKa = 7.91TFQVIQFYY331 pKa = 11.04QNNCKK336 pKa = 8.5VTGVAMHH343 pKa = 7.38AIGDD347 pKa = 3.88EE348 pKa = 4.31STPASYY354 pKa = 10.94VSSVDD359 pKa = 3.11VDD361 pKa = 3.79FTLPYY366 pKa = 9.73NASSTAALGYY376 pKa = 9.19MDD378 pKa = 4.99EE379 pKa = 4.53SSATFVLTDD388 pKa = 4.49GINPVVIPTQGEE400 pKa = 4.42SLVGFGFYY408 pKa = 9.56ATRR411 pKa = 11.84SIAGSIYY418 pKa = 9.24YY419 pKa = 7.91TAQSTQMALGLRR431 pKa = 11.84DD432 pKa = 3.6KK433 pKa = 11.34VFGEE437 pKa = 4.38VPPDD441 pKa = 3.4SALLFILIDD450 pKa = 3.82TVLSIPVNYY459 pKa = 10.27LKK461 pKa = 10.73LYY463 pKa = 8.05PQGFFTSSHH472 pKa = 5.27VDD474 pKa = 3.16QPIVYY479 pKa = 9.97DD480 pKa = 3.36SRR482 pKa = 11.84RR483 pKa = 11.84YY484 pKa = 8.42RR485 pKa = 11.84LEE487 pKa = 3.64YY488 pKa = 10.35VGLIEE493 pKa = 4.41VTTTVPTKK501 pKa = 10.45IEE503 pKa = 4.18FEE505 pKa = 4.24THH507 pKa = 5.43KK508 pKa = 10.79LAMSRR513 pKa = 11.84SFDD516 pKa = 3.53HH517 pKa = 6.3KK518 pKa = 10.46KK519 pKa = 10.12KK520 pKa = 10.67RR521 pKa = 11.84SGFLSAAYY529 pKa = 9.71HH530 pKa = 6.2GEE532 pKa = 4.15SAAA535 pKa = 4.04
Molecular weight: 57.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KM75|A0A1L3KM75_9VIRU Calici_coat domain-containing protein OS=Wuhan arthropod virus 3 OX=1923692 PE=4 SV=1
MM1 pKa = 7.43GVLSLATKK9 pKa = 9.32VGKK12 pKa = 9.9VAAKK16 pKa = 10.27GATTVNRR23 pKa = 11.84VVVNNGLPNTEE34 pKa = 3.52KK35 pKa = 10.64RR36 pKa = 11.84ISRR39 pKa = 11.84RR40 pKa = 11.84AARR43 pKa = 11.84YY44 pKa = 7.17EE45 pKa = 3.65RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.67AKK50 pKa = 10.58GKK52 pKa = 8.55FTTNLGAKK60 pKa = 8.53SAHH63 pKa = 5.62YY64 pKa = 10.45KK65 pKa = 10.31EE66 pKa = 5.55GIDD69 pKa = 3.25IAKK72 pKa = 8.94TKK74 pKa = 11.0GEE76 pKa = 3.99ASVFSQKK83 pKa = 10.22YY84 pKa = 9.3KK85 pKa = 10.41KK86 pKa = 9.99AAPKK90 pKa = 9.85KK91 pKa = 10.17APNKK95 pKa = 10.24HH96 pKa = 5.09LVKK99 pKa = 9.61TKK101 pKa = 10.61KK102 pKa = 9.6VVKK105 pKa = 10.0PITKK109 pKa = 8.25VTNPKK114 pKa = 10.38SLLKK118 pKa = 9.63PGNMGVAKK126 pKa = 10.38AITKK130 pKa = 8.91AVPVKK135 pKa = 10.47SVAKK139 pKa = 9.05VAPVKK144 pKa = 10.39PLTKK148 pKa = 10.06VASSTTKK155 pKa = 10.28KK156 pKa = 10.61SVVKK160 pKa = 10.73APIQSNSKK168 pKa = 10.38FDD170 pKa = 4.42DD171 pKa = 4.01PLKK174 pKa = 10.9KK175 pKa = 10.46GFPSNSTGAAGVKK188 pKa = 9.75PPPPKK193 pKa = 10.46DD194 pKa = 3.26LGLVKK199 pKa = 8.92PTPAKK204 pKa = 10.38VLSSATKK211 pKa = 9.97EE212 pKa = 3.98KK213 pKa = 9.97FTIPRR218 pKa = 11.84PTLVTKK224 pKa = 10.29AKK226 pKa = 10.04PVAKK230 pKa = 8.51PTAVVPPTKK239 pKa = 10.22VVKK242 pKa = 10.14PPAKK246 pKa = 10.15AGTPTTPAKK255 pKa = 10.97VNTPVKK261 pKa = 10.26KK262 pKa = 9.4ITDD265 pKa = 3.66VVPSKK270 pKa = 10.58GTVGKK275 pKa = 10.14IDD277 pKa = 4.88DD278 pKa = 4.49LTPKK282 pKa = 10.34PKK284 pKa = 10.55LKK286 pKa = 10.66GMAKK290 pKa = 9.76YY291 pKa = 9.46QALAKK296 pKa = 8.82KK297 pKa = 9.56HH298 pKa = 4.9KK299 pKa = 9.13WKK301 pKa = 9.98IAKK304 pKa = 9.74AVGGGTLSAVGATAMGLLGYY324 pKa = 7.56QAKK327 pKa = 9.71VAAANIAKK335 pKa = 10.13NSNRR339 pKa = 11.84EE340 pKa = 3.91TLIYY344 pKa = 10.68DD345 pKa = 3.54ATKK348 pKa = 10.83SSAKK352 pKa = 9.98YY353 pKa = 10.14AINEE357 pKa = 4.06KK358 pKa = 10.85EE359 pKa = 4.08EE360 pKa = 3.96AQNAAKK366 pKa = 10.17AQKK369 pKa = 9.72QRR371 pKa = 11.84EE372 pKa = 3.94EE373 pKa = 4.37DD374 pKa = 3.56LANTATTNKK383 pKa = 9.73HH384 pKa = 5.75LEE386 pKa = 4.51AIANKK391 pKa = 7.8PTLASMLTTKK401 pKa = 10.15RR402 pKa = 11.84QKK404 pKa = 11.16SNIRR408 pKa = 11.84YY409 pKa = 8.68QVGNNQVSRR418 pKa = 11.84NPLRR422 pKa = 11.84KK423 pKa = 8.48VTNSRR428 pKa = 11.84PQIDD432 pKa = 3.7KK433 pKa = 10.65LAPEE437 pKa = 4.41PSFLDD442 pKa = 3.36SLRR445 pKa = 11.84FKK447 pKa = 11.53SNMVV451 pKa = 3.13
MM1 pKa = 7.43GVLSLATKK9 pKa = 9.32VGKK12 pKa = 9.9VAAKK16 pKa = 10.27GATTVNRR23 pKa = 11.84VVVNNGLPNTEE34 pKa = 3.52KK35 pKa = 10.64RR36 pKa = 11.84ISRR39 pKa = 11.84RR40 pKa = 11.84AARR43 pKa = 11.84YY44 pKa = 7.17EE45 pKa = 3.65RR46 pKa = 11.84RR47 pKa = 11.84KK48 pKa = 10.67AKK50 pKa = 10.58GKK52 pKa = 8.55FTTNLGAKK60 pKa = 8.53SAHH63 pKa = 5.62YY64 pKa = 10.45KK65 pKa = 10.31EE66 pKa = 5.55GIDD69 pKa = 3.25IAKK72 pKa = 8.94TKK74 pKa = 11.0GEE76 pKa = 3.99ASVFSQKK83 pKa = 10.22YY84 pKa = 9.3KK85 pKa = 10.41KK86 pKa = 9.99AAPKK90 pKa = 9.85KK91 pKa = 10.17APNKK95 pKa = 10.24HH96 pKa = 5.09LVKK99 pKa = 9.61TKK101 pKa = 10.61KK102 pKa = 9.6VVKK105 pKa = 10.0PITKK109 pKa = 8.25VTNPKK114 pKa = 10.38SLLKK118 pKa = 9.63PGNMGVAKK126 pKa = 10.38AITKK130 pKa = 8.91AVPVKK135 pKa = 10.47SVAKK139 pKa = 9.05VAPVKK144 pKa = 10.39PLTKK148 pKa = 10.06VASSTTKK155 pKa = 10.28KK156 pKa = 10.61SVVKK160 pKa = 10.73APIQSNSKK168 pKa = 10.38FDD170 pKa = 4.42DD171 pKa = 4.01PLKK174 pKa = 10.9KK175 pKa = 10.46GFPSNSTGAAGVKK188 pKa = 9.75PPPPKK193 pKa = 10.46DD194 pKa = 3.26LGLVKK199 pKa = 8.92PTPAKK204 pKa = 10.38VLSSATKK211 pKa = 9.97EE212 pKa = 3.98KK213 pKa = 9.97FTIPRR218 pKa = 11.84PTLVTKK224 pKa = 10.29AKK226 pKa = 10.04PVAKK230 pKa = 8.51PTAVVPPTKK239 pKa = 10.22VVKK242 pKa = 10.14PPAKK246 pKa = 10.15AGTPTTPAKK255 pKa = 10.97VNTPVKK261 pKa = 10.26KK262 pKa = 9.4ITDD265 pKa = 3.66VVPSKK270 pKa = 10.58GTVGKK275 pKa = 10.14IDD277 pKa = 4.88DD278 pKa = 4.49LTPKK282 pKa = 10.34PKK284 pKa = 10.55LKK286 pKa = 10.66GMAKK290 pKa = 9.76YY291 pKa = 9.46QALAKK296 pKa = 8.82KK297 pKa = 9.56HH298 pKa = 4.9KK299 pKa = 9.13WKK301 pKa = 9.98IAKK304 pKa = 9.74AVGGGTLSAVGATAMGLLGYY324 pKa = 7.56QAKK327 pKa = 9.71VAAANIAKK335 pKa = 10.13NSNRR339 pKa = 11.84EE340 pKa = 3.91TLIYY344 pKa = 10.68DD345 pKa = 3.54ATKK348 pKa = 10.83SSAKK352 pKa = 9.98YY353 pKa = 10.14AINEE357 pKa = 4.06KK358 pKa = 10.85EE359 pKa = 4.08EE360 pKa = 3.96AQNAAKK366 pKa = 10.17AQKK369 pKa = 9.72QRR371 pKa = 11.84EE372 pKa = 3.94EE373 pKa = 4.37DD374 pKa = 3.56LANTATTNKK383 pKa = 9.73HH384 pKa = 5.75LEE386 pKa = 4.51AIANKK391 pKa = 7.8PTLASMLTTKK401 pKa = 10.15RR402 pKa = 11.84QKK404 pKa = 11.16SNIRR408 pKa = 11.84YY409 pKa = 8.68QVGNNQVSRR418 pKa = 11.84NPLRR422 pKa = 11.84KK423 pKa = 8.48VTNSRR428 pKa = 11.84PQIDD432 pKa = 3.7KK433 pKa = 10.65LAPEE437 pKa = 4.41PSFLDD442 pKa = 3.36SLRR445 pKa = 11.84FKK447 pKa = 11.53SNMVV451 pKa = 3.13
Molecular weight: 48.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3229 |
451 |
2243 |
1076.3 |
119.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.773 ± 1.931 | 1.61 ± 0.661 |
6.039 ± 1.27 | 5.048 ± 1.019 |
4.614 ± 0.813 | 5.915 ± 0.329 |
2.137 ± 0.364 | 5.141 ± 0.424 |
7.371 ± 2.516 | 8.455 ± 0.829 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.449 | 4.645 ± 0.255 |
6.38 ± 0.505 | 3.314 ± 0.331 |
3.716 ± 0.427 | 6.906 ± 0.358 |
6.689 ± 1.441 | 6.999 ± 0.69 |
1.022 ± 0.383 | 3.964 ± 0.581 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
