
White clover cryptic virus 1 (isolate Boccardo/2004) (WCCV-1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Alphapartitivirus; White clover cryptic virus 1
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
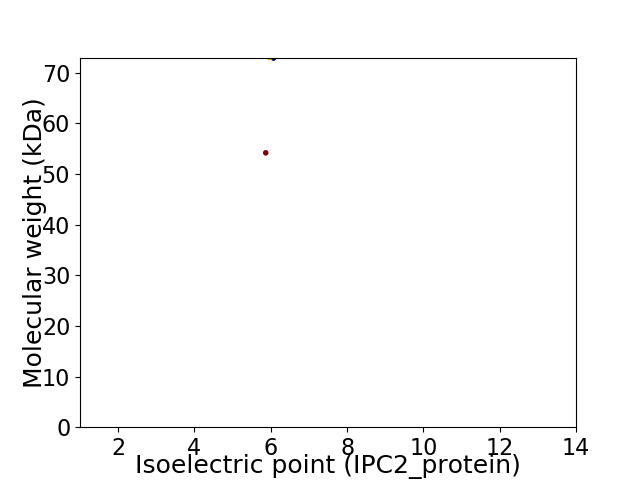
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q64FP0|RDRP_WCCVB RNA-directed RNA polymerase OS=White clover cryptic virus 1 (isolate Boccardo/2004) OX=654934 PE=4 SV=1
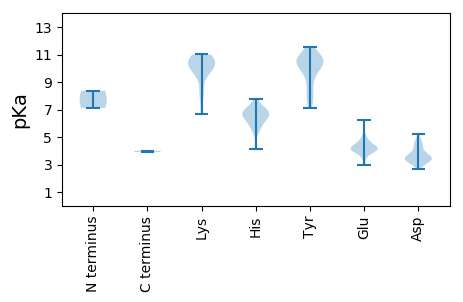
MM1 pKa = 7.12NQDD4 pKa = 3.04TPLANLNGPEE14 pKa = 4.17VPSGNVPPANPPGRR28 pKa = 11.84TNVAPPAQGAVQQPPAPAARR48 pKa = 11.84RR49 pKa = 11.84ARR51 pKa = 11.84NPHH54 pKa = 6.1GPVPPAGPSRR64 pKa = 11.84SAGAPAMLEE73 pKa = 3.92LSAAYY78 pKa = 9.4PMYY81 pKa = 9.88TEE83 pKa = 4.08QRR85 pKa = 11.84RR86 pKa = 11.84APNYY90 pKa = 8.66YY91 pKa = 10.39VPDD94 pKa = 3.96AQLLFHH100 pKa = 6.59TLGVCDD106 pKa = 4.23QLMTTTDD113 pKa = 3.01RR114 pKa = 11.84FLRR117 pKa = 11.84SMPSWLPIVSQLYY130 pKa = 10.48VSVLWNVMILKK141 pKa = 10.19VYY143 pKa = 10.81VNTGYY148 pKa = 10.8GAAYY152 pKa = 10.27AHH154 pKa = 7.04DD155 pKa = 5.25LDD157 pKa = 4.43VLLNHH162 pKa = 6.57LQINEE167 pKa = 3.97CMIPGPLVPFFQSLAAVNGPFDD189 pKa = 4.6WIGDD193 pKa = 4.56IIPAMPSFTEE203 pKa = 3.98LWTDD207 pKa = 3.29EE208 pKa = 4.49FAPHH212 pKa = 6.54AAYY215 pKa = 10.1ARR217 pKa = 11.84QIPIPAILLDD227 pKa = 3.38QLYY230 pKa = 10.04RR231 pKa = 11.84FATLAFDD238 pKa = 4.78AQLQTNYY245 pKa = 9.32ATFEE249 pKa = 3.98WYY251 pKa = 10.88SNIFNQDD258 pKa = 3.01VNTHH262 pKa = 5.36NARR265 pKa = 11.84LRR267 pKa = 11.84LGPQLCGSLFTTQAQCDD284 pKa = 3.79SARR287 pKa = 11.84AFWNPAFANGFTRR300 pKa = 11.84IDD302 pKa = 3.56AANGPLMAFPQLLGFISQDD321 pKa = 3.29GALQSNWFMHH331 pKa = 6.72ISLIMHH337 pKa = 7.35KK338 pKa = 10.0YY339 pKa = 8.1AQYY342 pKa = 11.07FNGSVPLKK350 pKa = 10.23SISPVGIGASVIYY363 pKa = 7.38GTPLEE368 pKa = 4.45DD369 pKa = 3.29TNVRR373 pKa = 11.84DD374 pKa = 3.6WLYY377 pKa = 10.25PAAAAIAPFRR387 pKa = 11.84STRR390 pKa = 11.84FLPRR394 pKa = 11.84RR395 pKa = 11.84EE396 pKa = 4.25LPATLAVRR404 pKa = 11.84FAHH407 pKa = 6.83ADD409 pKa = 3.46HH410 pKa = 6.93EE411 pKa = 4.45IEE413 pKa = 4.27EE414 pKa = 4.22QAEE417 pKa = 4.28QYY419 pKa = 11.08SILCHH424 pKa = 6.29TNMKK428 pKa = 9.62WYY430 pKa = 10.66VNNATQNNHH439 pKa = 4.48TAIEE443 pKa = 4.25GNYY446 pKa = 8.51IHH448 pKa = 6.78QGEE451 pKa = 4.68YY452 pKa = 9.67WNFTPFRR459 pKa = 11.84YY460 pKa = 9.92SPPVSLKK467 pKa = 8.41TQFAQVIASRR477 pKa = 11.84YY478 pKa = 7.5HH479 pKa = 4.14QQAANRR485 pKa = 11.84AEE487 pKa = 3.91
MM1 pKa = 7.12NQDD4 pKa = 3.04TPLANLNGPEE14 pKa = 4.17VPSGNVPPANPPGRR28 pKa = 11.84TNVAPPAQGAVQQPPAPAARR48 pKa = 11.84RR49 pKa = 11.84ARR51 pKa = 11.84NPHH54 pKa = 6.1GPVPPAGPSRR64 pKa = 11.84SAGAPAMLEE73 pKa = 3.92LSAAYY78 pKa = 9.4PMYY81 pKa = 9.88TEE83 pKa = 4.08QRR85 pKa = 11.84RR86 pKa = 11.84APNYY90 pKa = 8.66YY91 pKa = 10.39VPDD94 pKa = 3.96AQLLFHH100 pKa = 6.59TLGVCDD106 pKa = 4.23QLMTTTDD113 pKa = 3.01RR114 pKa = 11.84FLRR117 pKa = 11.84SMPSWLPIVSQLYY130 pKa = 10.48VSVLWNVMILKK141 pKa = 10.19VYY143 pKa = 10.81VNTGYY148 pKa = 10.8GAAYY152 pKa = 10.27AHH154 pKa = 7.04DD155 pKa = 5.25LDD157 pKa = 4.43VLLNHH162 pKa = 6.57LQINEE167 pKa = 3.97CMIPGPLVPFFQSLAAVNGPFDD189 pKa = 4.6WIGDD193 pKa = 4.56IIPAMPSFTEE203 pKa = 3.98LWTDD207 pKa = 3.29EE208 pKa = 4.49FAPHH212 pKa = 6.54AAYY215 pKa = 10.1ARR217 pKa = 11.84QIPIPAILLDD227 pKa = 3.38QLYY230 pKa = 10.04RR231 pKa = 11.84FATLAFDD238 pKa = 4.78AQLQTNYY245 pKa = 9.32ATFEE249 pKa = 3.98WYY251 pKa = 10.88SNIFNQDD258 pKa = 3.01VNTHH262 pKa = 5.36NARR265 pKa = 11.84LRR267 pKa = 11.84LGPQLCGSLFTTQAQCDD284 pKa = 3.79SARR287 pKa = 11.84AFWNPAFANGFTRR300 pKa = 11.84IDD302 pKa = 3.56AANGPLMAFPQLLGFISQDD321 pKa = 3.29GALQSNWFMHH331 pKa = 6.72ISLIMHH337 pKa = 7.35KK338 pKa = 10.0YY339 pKa = 8.1AQYY342 pKa = 11.07FNGSVPLKK350 pKa = 10.23SISPVGIGASVIYY363 pKa = 7.38GTPLEE368 pKa = 4.45DD369 pKa = 3.29TNVRR373 pKa = 11.84DD374 pKa = 3.6WLYY377 pKa = 10.25PAAAAIAPFRR387 pKa = 11.84STRR390 pKa = 11.84FLPRR394 pKa = 11.84RR395 pKa = 11.84EE396 pKa = 4.25LPATLAVRR404 pKa = 11.84FAHH407 pKa = 6.83ADD409 pKa = 3.46HH410 pKa = 6.93EE411 pKa = 4.45IEE413 pKa = 4.27EE414 pKa = 4.22QAEE417 pKa = 4.28QYY419 pKa = 11.08SILCHH424 pKa = 6.29TNMKK428 pKa = 9.62WYY430 pKa = 10.66VNNATQNNHH439 pKa = 4.48TAIEE443 pKa = 4.25GNYY446 pKa = 8.51IHH448 pKa = 6.78QGEE451 pKa = 4.68YY452 pKa = 9.67WNFTPFRR459 pKa = 11.84YY460 pKa = 9.92SPPVSLKK467 pKa = 8.41TQFAQVIASRR477 pKa = 11.84YY478 pKa = 7.5HH479 pKa = 4.14QQAANRR485 pKa = 11.84AEE487 pKa = 3.91
Molecular weight: 54.16 kDa
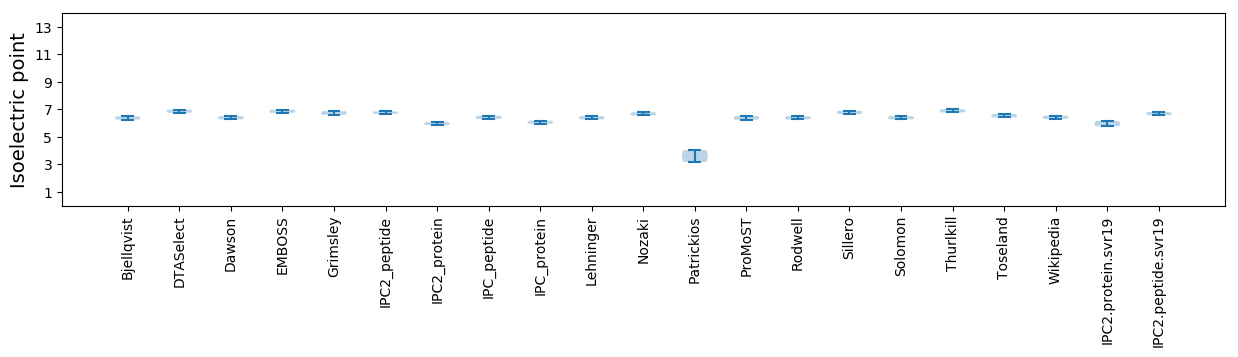
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q64FP0|RDRP_WCCVB RNA-directed RNA polymerase OS=White clover cryptic virus 1 (isolate Boccardo/2004) OX=654934 PE=4 SV=1
MM1 pKa = 8.36DD2 pKa = 4.02YY3 pKa = 11.07LITAFNRR10 pKa = 11.84ITHH13 pKa = 6.73WFLTPTNLEE22 pKa = 4.34YY23 pKa = 10.35IGSYY27 pKa = 10.09SLPPGLLRR35 pKa = 11.84VNDD38 pKa = 3.61VAVANHH44 pKa = 6.96KK45 pKa = 9.18ATLDD49 pKa = 3.52RR50 pKa = 11.84SFDD53 pKa = 3.3KK54 pKa = 11.0YY55 pKa = 11.08LYY57 pKa = 8.91EE58 pKa = 5.05HH59 pKa = 7.76EE60 pKa = 4.75INLITKK66 pKa = 7.7EE67 pKa = 4.0YY68 pKa = 9.45RR69 pKa = 11.84RR70 pKa = 11.84SPIDD74 pKa = 3.37EE75 pKa = 4.61DD76 pKa = 4.94SILEE80 pKa = 4.22DD81 pKa = 4.41FFSGDD86 pKa = 3.14LPYY89 pKa = 10.76FEE91 pKa = 5.74IPFDD95 pKa = 3.56EE96 pKa = 4.49HH97 pKa = 7.54VEE99 pKa = 4.07RR100 pKa = 11.84GLEE103 pKa = 3.88CMAAAFRR110 pKa = 11.84PPRR113 pKa = 11.84PCRR116 pKa = 11.84PAHH119 pKa = 6.19ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.36PPFSTDD142 pKa = 3.4EE143 pKa = 4.16YY144 pKa = 10.97FLSQRR149 pKa = 11.84KK150 pKa = 6.69TFGEE154 pKa = 5.0FIRR157 pKa = 11.84MHH159 pKa = 6.57EE160 pKa = 4.18YY161 pKa = 10.58EE162 pKa = 5.75HH163 pKa = 7.05IDD165 pKa = 3.17KK166 pKa = 10.9EE167 pKa = 4.62DD168 pKa = 3.31FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 5.9PNIEE177 pKa = 3.71SHH179 pKa = 7.13DD180 pKa = 3.9FLRR183 pKa = 11.84TVVPPKK189 pKa = 10.59FGFLKK194 pKa = 11.0SMIFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.51HH206 pKa = 6.39IIKK209 pKa = 10.57SGFQDD214 pKa = 3.54STDD217 pKa = 3.88LEE219 pKa = 4.27QTGYY223 pKa = 10.41FFNRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.3TAIVKK242 pKa = 10.57KK243 pKa = 10.26NDD245 pKa = 3.58PNKK248 pKa = 9.91MRR250 pKa = 11.84TIWGASKK257 pKa = 9.78PWIIAEE263 pKa = 4.1TMFYY267 pKa = 10.21WEE269 pKa = 3.66YY270 pKa = 10.78LAWIKK275 pKa = 10.55HH276 pKa = 5.35NPGATPMLWGYY287 pKa = 7.52EE288 pKa = 4.17TFTGGWFRR296 pKa = 11.84LNHH299 pKa = 6.01EE300 pKa = 4.88LFCGLIQRR308 pKa = 11.84SFLTLDD314 pKa = 3.43WSRR317 pKa = 11.84FDD319 pKa = 3.15KK320 pKa = 10.78RR321 pKa = 11.84AYY323 pKa = 10.14FPLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 7.1TVKK335 pKa = 9.88TFLTFEE341 pKa = 4.03EE342 pKa = 6.24GYY344 pKa = 10.49VPTHH348 pKa = 6.99AAPTHH353 pKa = 5.51PQWSQEE359 pKa = 4.05NIDD362 pKa = 3.57RR363 pKa = 11.84LEE365 pKa = 4.18RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.3TMLATILSALHH422 pKa = 6.9FDD424 pKa = 4.25PLHH427 pKa = 6.76CIIKK431 pKa = 9.93VQGDD435 pKa = 3.68DD436 pKa = 3.67SILRR440 pKa = 11.84LTTLIPVDD448 pKa = 3.23QHH450 pKa = 5.67TNFMDD455 pKa = 4.66HH456 pKa = 6.98IVRR459 pKa = 11.84LADD462 pKa = 3.4TYY464 pKa = 11.33FNSIVNVKK472 pKa = 10.09KK473 pKa = 10.93SEE475 pKa = 3.85VRR477 pKa = 11.84NSLNGCEE484 pKa = 3.92VLSYY488 pKa = 10.62RR489 pKa = 11.84NHH491 pKa = 6.48NGLPHH496 pKa = 6.86RR497 pKa = 11.84DD498 pKa = 4.22EE499 pKa = 4.25ITMLAQFYY507 pKa = 7.85HH508 pKa = 6.12TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.24ASCANHH534 pKa = 6.33NRR536 pKa = 11.84VLWVLEE542 pKa = 3.86DD543 pKa = 3.44VYY545 pKa = 11.58NYY547 pKa = 10.55YY548 pKa = 10.35RR549 pKa = 11.84DD550 pKa = 3.28LGYY553 pKa = 10.61RR554 pKa = 11.84PNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.55SPDD569 pKa = 3.29LTMPEE574 pKa = 4.14MPLDD578 pKa = 3.84HH579 pKa = 7.26FPTKK583 pKa = 10.6SEE585 pKa = 2.97IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 8.33HH590 pKa = 6.22TEE592 pKa = 3.07THH594 pKa = 5.74YY595 pKa = 11.25QNEE598 pKa = 4.3AQNARR603 pKa = 11.84TWPRR607 pKa = 11.84TLFINAPGEE616 pKa = 4.0
MM1 pKa = 8.36DD2 pKa = 4.02YY3 pKa = 11.07LITAFNRR10 pKa = 11.84ITHH13 pKa = 6.73WFLTPTNLEE22 pKa = 4.34YY23 pKa = 10.35IGSYY27 pKa = 10.09SLPPGLLRR35 pKa = 11.84VNDD38 pKa = 3.61VAVANHH44 pKa = 6.96KK45 pKa = 9.18ATLDD49 pKa = 3.52RR50 pKa = 11.84SFDD53 pKa = 3.3KK54 pKa = 11.0YY55 pKa = 11.08LYY57 pKa = 8.91EE58 pKa = 5.05HH59 pKa = 7.76EE60 pKa = 4.75INLITKK66 pKa = 7.7EE67 pKa = 4.0YY68 pKa = 9.45RR69 pKa = 11.84RR70 pKa = 11.84SPIDD74 pKa = 3.37EE75 pKa = 4.61DD76 pKa = 4.94SILEE80 pKa = 4.22DD81 pKa = 4.41FFSGDD86 pKa = 3.14LPYY89 pKa = 10.76FEE91 pKa = 5.74IPFDD95 pKa = 3.56EE96 pKa = 4.49HH97 pKa = 7.54VEE99 pKa = 4.07RR100 pKa = 11.84GLEE103 pKa = 3.88CMAAAFRR110 pKa = 11.84PPRR113 pKa = 11.84PCRR116 pKa = 11.84PAHH119 pKa = 6.19ILDD122 pKa = 3.78VKK124 pKa = 10.61HH125 pKa = 6.44GYY127 pKa = 8.11PYY129 pKa = 10.43KK130 pKa = 10.06WNVNAEE136 pKa = 4.36PPFSTDD142 pKa = 3.4EE143 pKa = 4.16YY144 pKa = 10.97FLSQRR149 pKa = 11.84KK150 pKa = 6.69TFGEE154 pKa = 5.0FIRR157 pKa = 11.84MHH159 pKa = 6.57EE160 pKa = 4.18YY161 pKa = 10.58EE162 pKa = 5.75HH163 pKa = 7.05IDD165 pKa = 3.17KK166 pKa = 10.9EE167 pKa = 4.62DD168 pKa = 3.31FFRR171 pKa = 11.84RR172 pKa = 11.84HH173 pKa = 5.9PNIEE177 pKa = 3.71SHH179 pKa = 7.13DD180 pKa = 3.9FLRR183 pKa = 11.84TVVPPKK189 pKa = 10.59FGFLKK194 pKa = 11.0SMIFSWTRR202 pKa = 11.84RR203 pKa = 11.84WHH205 pKa = 6.51HH206 pKa = 6.39IIKK209 pKa = 10.57SGFQDD214 pKa = 3.54STDD217 pKa = 3.88LEE219 pKa = 4.27QTGYY223 pKa = 10.41FFNRR227 pKa = 11.84FIFPMLLHH235 pKa = 6.66TKK237 pKa = 8.3TAIVKK242 pKa = 10.57KK243 pKa = 10.26NDD245 pKa = 3.58PNKK248 pKa = 9.91MRR250 pKa = 11.84TIWGASKK257 pKa = 9.78PWIIAEE263 pKa = 4.1TMFYY267 pKa = 10.21WEE269 pKa = 3.66YY270 pKa = 10.78LAWIKK275 pKa = 10.55HH276 pKa = 5.35NPGATPMLWGYY287 pKa = 7.52EE288 pKa = 4.17TFTGGWFRR296 pKa = 11.84LNHH299 pKa = 6.01EE300 pKa = 4.88LFCGLIQRR308 pKa = 11.84SFLTLDD314 pKa = 3.43WSRR317 pKa = 11.84FDD319 pKa = 3.15KK320 pKa = 10.78RR321 pKa = 11.84AYY323 pKa = 10.14FPLLRR328 pKa = 11.84RR329 pKa = 11.84ILYY332 pKa = 7.1TVKK335 pKa = 9.88TFLTFEE341 pKa = 4.03EE342 pKa = 6.24GYY344 pKa = 10.49VPTHH348 pKa = 6.99AAPTHH353 pKa = 5.51PQWSQEE359 pKa = 4.05NIDD362 pKa = 3.57RR363 pKa = 11.84LEE365 pKa = 4.18RR366 pKa = 11.84LWLWTLEE373 pKa = 4.18NLFEE377 pKa = 4.94APIILPDD384 pKa = 2.89GRR386 pKa = 11.84MYY388 pKa = 10.65RR389 pKa = 11.84RR390 pKa = 11.84HH391 pKa = 5.78FAGIPSGLFITQLLDD406 pKa = 2.7SWYY409 pKa = 9.85NYY411 pKa = 9.3TMLATILSALHH422 pKa = 6.9FDD424 pKa = 4.25PLHH427 pKa = 6.76CIIKK431 pKa = 9.93VQGDD435 pKa = 3.68DD436 pKa = 3.67SILRR440 pKa = 11.84LTTLIPVDD448 pKa = 3.23QHH450 pKa = 5.67TNFMDD455 pKa = 4.66HH456 pKa = 6.98IVRR459 pKa = 11.84LADD462 pKa = 3.4TYY464 pKa = 11.33FNSIVNVKK472 pKa = 10.09KK473 pKa = 10.93SEE475 pKa = 3.85VRR477 pKa = 11.84NSLNGCEE484 pKa = 3.92VLSYY488 pKa = 10.62RR489 pKa = 11.84NHH491 pKa = 6.48NGLPHH496 pKa = 6.86RR497 pKa = 11.84DD498 pKa = 4.22EE499 pKa = 4.25ITMLAQFYY507 pKa = 7.85HH508 pKa = 6.12TKK510 pKa = 10.61ARR512 pKa = 11.84DD513 pKa = 3.58PTPEE517 pKa = 3.33ITMAQAIGFAYY528 pKa = 10.24ASCANHH534 pKa = 6.33NRR536 pKa = 11.84VLWVLEE542 pKa = 3.86DD543 pKa = 3.44VYY545 pKa = 11.58NYY547 pKa = 10.55YY548 pKa = 10.35RR549 pKa = 11.84DD550 pKa = 3.28LGYY553 pKa = 10.61RR554 pKa = 11.84PNRR557 pKa = 11.84AGLTLTFGDD566 pKa = 4.55SPDD569 pKa = 3.29LTMPEE574 pKa = 4.14MPLDD578 pKa = 3.84HH579 pKa = 7.26FPTKK583 pKa = 10.6SEE585 pKa = 2.97IRR587 pKa = 11.84RR588 pKa = 11.84YY589 pKa = 8.33HH590 pKa = 6.22TEE592 pKa = 3.07THH594 pKa = 5.74YY595 pKa = 11.25QNEE598 pKa = 4.3AQNARR603 pKa = 11.84TWPRR607 pKa = 11.84TLFINAPGEE616 pKa = 4.0
Molecular weight: 72.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103 |
487 |
616 |
551.5 |
63.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.522 ± 2.233 | 0.997 ± 0.016 |
4.805 ± 0.619 | 4.896 ± 0.898 |
6.165 ± 0.575 | 4.714 ± 0.234 |
3.898 ± 0.571 | 5.802 ± 0.488 |
2.539 ± 0.843 | 9.157 ± 0.297 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.448 ± 0.009 | 5.621 ± 0.53 |
7.797 ± 0.92 | 3.898 ± 1.261 |
6.074 ± 0.639 | 4.805 ± 0.183 |
6.437 ± 0.612 | 4.17 ± 0.537 |
2.448 ± 0.22 | 4.805 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
