
Allochromatium warmingii (Chromatium warmingii)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Chromatiaceae;
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
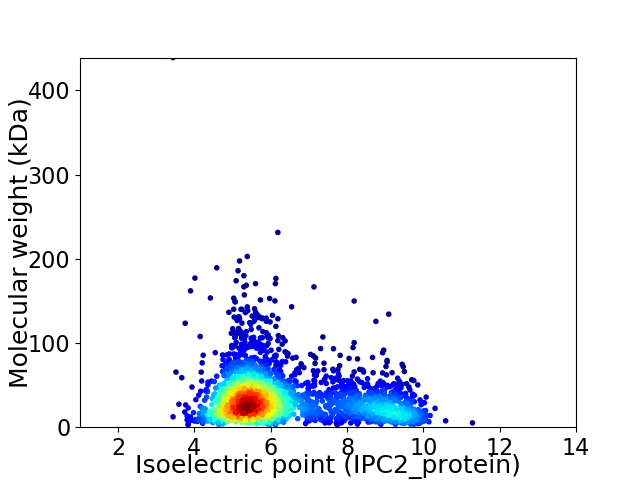
Virtual 2D-PAGE plot for 2689 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H3EAA2|A0A1H3EAA2_ALLWA Multifunctional CCA protein OS=Allochromatium warmingii OX=61595 GN=cca PE=3 SV=1
MM1 pKa = 7.17TVASDD6 pKa = 3.72LLEE9 pKa = 4.87GYY11 pKa = 10.32EE12 pKa = 4.55SIPVEE17 pKa = 4.18NRR19 pKa = 11.84NEE21 pKa = 3.78ATQQFIQILNEE32 pKa = 3.95MPEE35 pKa = 4.1DD36 pKa = 3.81ADD38 pKa = 3.9FVEE41 pKa = 6.07GPTNSQPGEE50 pKa = 3.98TSYY53 pKa = 12.06SLLPPPAQGEE63 pKa = 4.36TGQAEE68 pKa = 4.4GGDD71 pKa = 4.17GYY73 pKa = 11.14DD74 pKa = 5.14IIITDD79 pKa = 4.87LPPDD83 pKa = 3.85TEE85 pKa = 4.77SDD87 pKa = 3.55FTVANNVEE95 pKa = 3.97QFIILGSGSANLQQEE110 pKa = 4.49TPPPGNQQPTQFFTGNGDD128 pKa = 3.6DD129 pKa = 3.96SLLTGGAGSQGVFGGGDD146 pKa = 3.53DD147 pKa = 3.88TVTSATAVTGQNPTMSQMTLINGTAAAKK175 pKa = 8.78WWDD178 pKa = 3.51EE179 pKa = 3.76ADD181 pKa = 3.72YY182 pKa = 10.95LANNPDD188 pKa = 2.97VAQAVQNGALVSGYY202 pKa = 7.02QHH204 pKa = 6.84YY205 pKa = 10.7ILFGAAEE212 pKa = 4.25NRR214 pKa = 11.84DD215 pKa = 3.85STVSSWWNEE224 pKa = 3.37GYY226 pKa = 11.13YY227 pKa = 10.44LDD229 pKa = 4.86NNPDD233 pKa = 3.07VAQAIQAGILNSGYY247 pKa = 7.71QHH249 pKa = 6.65FALYY253 pKa = 9.96GAAEE257 pKa = 4.19NRR259 pKa = 11.84VSTVSTWWNEE269 pKa = 3.26TDD271 pKa = 3.76YY272 pKa = 11.68LNNNPDD278 pKa = 2.84VAQAVQNGLLASGYY292 pKa = 6.97QHH294 pKa = 6.88FAMFGASEE302 pKa = 4.07NRR304 pKa = 11.84NLIRR308 pKa = 11.84SWWDD312 pKa = 2.84EE313 pKa = 3.65EE314 pKa = 5.52LYY316 pKa = 10.98LKK318 pKa = 10.81EE319 pKa = 4.17NTDD322 pKa = 3.32VAQAVADD329 pKa = 4.2GVFASGYY336 pKa = 9.59HH337 pKa = 5.55HH338 pKa = 6.96FAMYY342 pKa = 10.05GAAEE346 pKa = 3.97QRR348 pKa = 11.84DD349 pKa = 3.54PNAYY353 pKa = 10.12FNAADD358 pKa = 3.7YY359 pKa = 10.67LAANGDD365 pKa = 3.12VDD367 pKa = 3.48IAVQIGIFKK376 pKa = 10.72SAFEE380 pKa = 4.7HH381 pKa = 5.04YY382 pKa = 9.54QKK384 pKa = 11.13YY385 pKa = 9.12GAAEE389 pKa = 4.1GRR391 pKa = 11.84TPSDD395 pKa = 3.3DD396 pKa = 4.16FNAEE400 pKa = 3.87NYY402 pKa = 9.87LDD404 pKa = 3.52RR405 pKa = 11.84WQDD408 pKa = 3.23VEE410 pKa = 4.85DD411 pKa = 3.91AVNNGSLNAGQHH423 pKa = 4.89AVFYY427 pKa = 9.89GATEE431 pKa = 4.12GRR433 pKa = 11.84VSNDD437 pKa = 2.25VDD439 pKa = 2.9IWEE442 pKa = 4.9GGAGNDD448 pKa = 4.1TLDD451 pKa = 4.47GGLGDD456 pKa = 5.69DD457 pKa = 5.11DD458 pKa = 4.25LTGGTGSDD466 pKa = 3.19TFVFRR471 pKa = 11.84ANSDD475 pKa = 2.95EE476 pKa = 5.16DD477 pKa = 3.68IIRR480 pKa = 11.84DD481 pKa = 3.71FDD483 pKa = 4.38FKK485 pKa = 10.98TSATDD490 pKa = 3.69LEE492 pKa = 4.85KK493 pKa = 10.83DD494 pKa = 3.69TIQTIGLNVTAEE506 pKa = 4.16SLVAGVTEE514 pKa = 4.54DD515 pKa = 3.84ANGHH519 pKa = 5.52AVVTLSDD526 pKa = 3.84GNTITLIGVKK536 pKa = 10.09AADD539 pKa = 3.52VTADD543 pKa = 3.61MFDD546 pKa = 3.33IQPP549 pKa = 3.51
MM1 pKa = 7.17TVASDD6 pKa = 3.72LLEE9 pKa = 4.87GYY11 pKa = 10.32EE12 pKa = 4.55SIPVEE17 pKa = 4.18NRR19 pKa = 11.84NEE21 pKa = 3.78ATQQFIQILNEE32 pKa = 3.95MPEE35 pKa = 4.1DD36 pKa = 3.81ADD38 pKa = 3.9FVEE41 pKa = 6.07GPTNSQPGEE50 pKa = 3.98TSYY53 pKa = 12.06SLLPPPAQGEE63 pKa = 4.36TGQAEE68 pKa = 4.4GGDD71 pKa = 4.17GYY73 pKa = 11.14DD74 pKa = 5.14IIITDD79 pKa = 4.87LPPDD83 pKa = 3.85TEE85 pKa = 4.77SDD87 pKa = 3.55FTVANNVEE95 pKa = 3.97QFIILGSGSANLQQEE110 pKa = 4.49TPPPGNQQPTQFFTGNGDD128 pKa = 3.6DD129 pKa = 3.96SLLTGGAGSQGVFGGGDD146 pKa = 3.53DD147 pKa = 3.88TVTSATAVTGQNPTMSQMTLINGTAAAKK175 pKa = 8.78WWDD178 pKa = 3.51EE179 pKa = 3.76ADD181 pKa = 3.72YY182 pKa = 10.95LANNPDD188 pKa = 2.97VAQAVQNGALVSGYY202 pKa = 7.02QHH204 pKa = 6.84YY205 pKa = 10.7ILFGAAEE212 pKa = 4.25NRR214 pKa = 11.84DD215 pKa = 3.85STVSSWWNEE224 pKa = 3.37GYY226 pKa = 11.13YY227 pKa = 10.44LDD229 pKa = 4.86NNPDD233 pKa = 3.07VAQAIQAGILNSGYY247 pKa = 7.71QHH249 pKa = 6.65FALYY253 pKa = 9.96GAAEE257 pKa = 4.19NRR259 pKa = 11.84VSTVSTWWNEE269 pKa = 3.26TDD271 pKa = 3.76YY272 pKa = 11.68LNNNPDD278 pKa = 2.84VAQAVQNGLLASGYY292 pKa = 6.97QHH294 pKa = 6.88FAMFGASEE302 pKa = 4.07NRR304 pKa = 11.84NLIRR308 pKa = 11.84SWWDD312 pKa = 2.84EE313 pKa = 3.65EE314 pKa = 5.52LYY316 pKa = 10.98LKK318 pKa = 10.81EE319 pKa = 4.17NTDD322 pKa = 3.32VAQAVADD329 pKa = 4.2GVFASGYY336 pKa = 9.59HH337 pKa = 5.55HH338 pKa = 6.96FAMYY342 pKa = 10.05GAAEE346 pKa = 3.97QRR348 pKa = 11.84DD349 pKa = 3.54PNAYY353 pKa = 10.12FNAADD358 pKa = 3.7YY359 pKa = 10.67LAANGDD365 pKa = 3.12VDD367 pKa = 3.48IAVQIGIFKK376 pKa = 10.72SAFEE380 pKa = 4.7HH381 pKa = 5.04YY382 pKa = 9.54QKK384 pKa = 11.13YY385 pKa = 9.12GAAEE389 pKa = 4.1GRR391 pKa = 11.84TPSDD395 pKa = 3.3DD396 pKa = 4.16FNAEE400 pKa = 3.87NYY402 pKa = 9.87LDD404 pKa = 3.52RR405 pKa = 11.84WQDD408 pKa = 3.23VEE410 pKa = 4.85DD411 pKa = 3.91AVNNGSLNAGQHH423 pKa = 4.89AVFYY427 pKa = 9.89GATEE431 pKa = 4.12GRR433 pKa = 11.84VSNDD437 pKa = 2.25VDD439 pKa = 2.9IWEE442 pKa = 4.9GGAGNDD448 pKa = 4.1TLDD451 pKa = 4.47GGLGDD456 pKa = 5.69DD457 pKa = 5.11DD458 pKa = 4.25LTGGTGSDD466 pKa = 3.19TFVFRR471 pKa = 11.84ANSDD475 pKa = 2.95EE476 pKa = 5.16DD477 pKa = 3.68IIRR480 pKa = 11.84DD481 pKa = 3.71FDD483 pKa = 4.38FKK485 pKa = 10.98TSATDD490 pKa = 3.69LEE492 pKa = 4.85KK493 pKa = 10.83DD494 pKa = 3.69TIQTIGLNVTAEE506 pKa = 4.16SLVAGVTEE514 pKa = 4.54DD515 pKa = 3.84ANGHH519 pKa = 5.52AVVTLSDD526 pKa = 3.84GNTITLIGVKK536 pKa = 10.09AADD539 pKa = 3.52VTADD543 pKa = 3.61MFDD546 pKa = 3.33IQPP549 pKa = 3.51
Molecular weight: 58.81 kDa
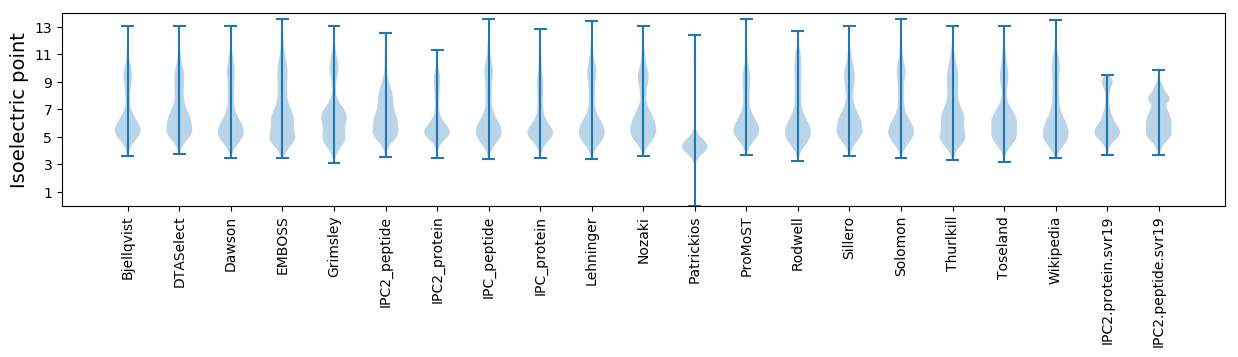
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H3BYQ7|A0A1H3BYQ7_ALLWA HD-like signal output (HDOD) domain no enzymatic activity OS=Allochromatium warmingii OX=61595 GN=SAMN05421644_10457 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.31VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.62GRR39 pKa = 11.84VRR41 pKa = 11.84LIPP44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LKK11 pKa = 10.48RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.93GFRR19 pKa = 11.84ARR21 pKa = 11.84SATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.31VLSARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.62GRR39 pKa = 11.84VRR41 pKa = 11.84LIPP44 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
889806 |
24 |
4262 |
330.9 |
36.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.627 ± 0.061 | 1.063 ± 0.019 |
5.646 ± 0.042 | 6.018 ± 0.041 |
3.405 ± 0.031 | 7.145 ± 0.048 |
2.468 ± 0.024 | 5.271 ± 0.036 |
2.748 ± 0.044 | 11.77 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.019 ± 0.025 | 2.78 ± 0.036 |
5.161 ± 0.042 | 4.398 ± 0.037 |
7.43 ± 0.05 | 5.027 ± 0.033 |
5.51 ± 0.044 | 6.702 ± 0.041 |
1.384 ± 0.022 | 2.427 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
