
Sphingomonas panacisoli
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
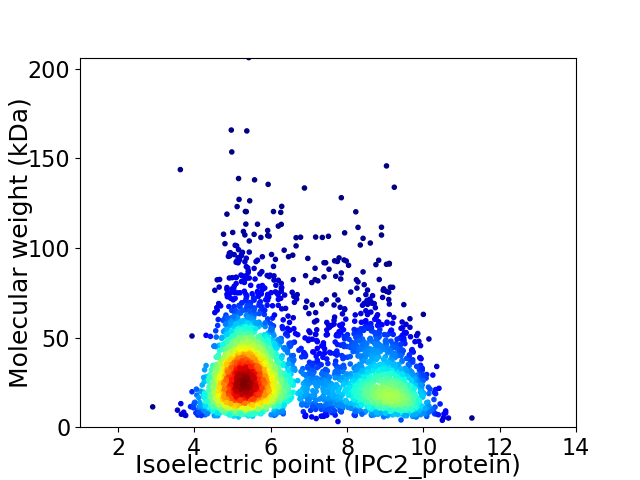
Virtual 2D-PAGE plot for 3225 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8LE43|A0A5B8LE43_9SPHN Glutathione S-transferase family protein OS=Sphingomonas panacisoli OX=1813879 GN=FPZ24_01650 PE=4 SV=1
MM1 pKa = 7.29TVPTEE6 pKa = 3.99PVTPEE11 pKa = 3.29MLLRR15 pKa = 11.84RR16 pKa = 11.84FLYY19 pKa = 10.23GVNEE23 pKa = 4.28TPDD26 pKa = 3.79LLTDD30 pKa = 4.15RR31 pKa = 11.84LRR33 pKa = 11.84DD34 pKa = 3.61LAPTASISIGEE45 pKa = 4.38INSDD49 pKa = 3.82TYY51 pKa = 10.38LASYY55 pKa = 9.75GRR57 pKa = 11.84YY58 pKa = 9.12INAAYY63 pKa = 9.93FPFVQDD69 pKa = 3.9FFTGAKK75 pKa = 7.66TLSVGTHH82 pKa = 5.52TVAQLQSTYY91 pKa = 10.62GASEE95 pKa = 3.91FLYY98 pKa = 10.38TLSNANTDD106 pKa = 3.3VGSSDD111 pKa = 3.32FFLRR115 pKa = 11.84AFMFGHH121 pKa = 7.43ADD123 pKa = 3.43FHH125 pKa = 7.01LSPDD129 pKa = 3.49TQFVVDD135 pKa = 4.07TNGDD139 pKa = 3.5LHH141 pKa = 8.22IEE143 pKa = 4.02NMRR146 pKa = 11.84YY147 pKa = 9.49IPDD150 pKa = 3.25VDD152 pKa = 4.52NFDD155 pKa = 4.31FDD157 pKa = 4.11STGPTAIYY165 pKa = 10.16DD166 pKa = 3.71AVFAKK171 pKa = 10.67PEE173 pKa = 4.17IDD175 pKa = 3.4PSSIGRR181 pKa = 11.84QITWTYY187 pKa = 10.02SDD189 pKa = 3.58PTIVHH194 pKa = 6.2NGYY197 pKa = 9.58YY198 pKa = 9.25AANYY202 pKa = 8.0AANVLNNYY210 pKa = 9.82NITSNSIYY218 pKa = 10.42AGAIILSQSFGYY230 pKa = 9.66FNQWKK235 pKa = 10.38SEE237 pKa = 3.61GVYY240 pKa = 10.95EE241 pKa = 4.43NFRR244 pKa = 11.84DD245 pKa = 3.89NKK247 pKa = 9.5TIIYY251 pKa = 9.81DD252 pKa = 3.7GLGTNVVVADD262 pKa = 3.49ITSLHH267 pKa = 6.03GSILIGGDD275 pKa = 3.78GNDD278 pKa = 3.63QLIGAHH284 pKa = 6.27YY285 pKa = 10.97DD286 pKa = 3.37DD287 pKa = 4.71VFYY290 pKa = 11.2GGDD293 pKa = 3.59GNDD296 pKa = 3.77TIKK299 pKa = 10.78PGLGSSEE306 pKa = 3.82LWGGDD311 pKa = 3.43EE312 pKa = 4.11EE313 pKa = 5.28SKK315 pKa = 10.99PEE317 pKa = 4.3YY318 pKa = 10.3EE319 pKa = 4.6DD320 pKa = 3.98TDD322 pKa = 3.73TADD325 pKa = 3.71YY326 pKa = 9.26TGHH329 pKa = 6.07SAIDD333 pKa = 3.27ITFDD337 pKa = 3.25NSGEE341 pKa = 4.07KK342 pKa = 10.53LSLKK346 pKa = 9.44VAHH349 pKa = 7.21DD350 pKa = 3.87GANDD354 pKa = 3.75TLHH357 pKa = 7.12SIEE360 pKa = 4.62VIKK363 pKa = 9.32GTPATDD369 pKa = 3.11HH370 pKa = 6.13VKK372 pKa = 10.46IIGSIPTATMLTIDD386 pKa = 3.69AAGGQGPNPLDD397 pKa = 3.94TINLAKK403 pKa = 10.13ADD405 pKa = 3.92NGIHH409 pKa = 6.42VSIANDD415 pKa = 3.56GTGYY419 pKa = 9.9IHH421 pKa = 8.11DD422 pKa = 4.49KK423 pKa = 8.03TTEE426 pKa = 3.81GDD428 pKa = 3.14ILLAGFNTQIIGSAFDD444 pKa = 5.37DD445 pKa = 4.18VFDD448 pKa = 5.91DD449 pKa = 4.21DD450 pKa = 5.35AVGAKK455 pKa = 10.11HH456 pKa = 6.95IDD458 pKa = 3.19GGYY461 pKa = 10.75GNDD464 pKa = 3.85TISITGASDD473 pKa = 4.05DD474 pKa = 3.92SLLLGGAGNDD484 pKa = 4.14TITGGDD490 pKa = 3.77GNDD493 pKa = 3.46VLVGGAYY500 pKa = 9.99AYY502 pKa = 9.97GAYY505 pKa = 10.24SDD507 pKa = 4.06TLNGGGGNDD516 pKa = 3.48MLIGSGANDD525 pKa = 3.44IFDD528 pKa = 4.29GGDD531 pKa = 3.28GYY533 pKa = 11.47DD534 pKa = 3.89YY535 pKa = 10.8IRR537 pKa = 11.84PTAIPGTTFGLATITGGAGDD557 pKa = 4.16DD558 pKa = 3.98VINLRR563 pKa = 11.84EE564 pKa = 4.44GGDD567 pKa = 3.76HH568 pKa = 6.08YY569 pKa = 11.72NGMYY573 pKa = 10.06HH574 pKa = 6.63YY575 pKa = 10.76GAAPTLNFGANDD587 pKa = 3.43GHH589 pKa = 6.44DD590 pKa = 3.83TVLTASGGDD599 pKa = 3.35VFTAYY604 pKa = 10.76DD605 pKa = 3.53NGVLNDD611 pKa = 4.15HH612 pKa = 6.54GADD615 pKa = 4.18GIVVKK620 pKa = 10.91LDD622 pKa = 3.11GLYY625 pKa = 10.85ASDD628 pKa = 5.37IDD630 pKa = 4.68IIWDD634 pKa = 3.42VTPQPEE640 pKa = 4.39PFFGLGFNGNGDD652 pKa = 3.7LAIVVKK658 pKa = 9.88STGASIFIEE667 pKa = 4.42DD668 pKa = 3.88VNGSATFGNQSSSSNHH684 pKa = 5.9FGVTILDD691 pKa = 3.84GAGNEE696 pKa = 3.43IWIPSFPVQYY706 pKa = 11.03GDD708 pKa = 3.26LSSYY712 pKa = 10.71RR713 pKa = 11.84GGEE716 pKa = 3.53AAYY719 pKa = 10.17FDD721 pKa = 4.23AAALSSSEE729 pKa = 4.17STGTSGDD736 pKa = 3.5DD737 pKa = 3.38VMSGGRR743 pKa = 11.84GDD745 pKa = 5.3DD746 pKa = 4.01DD747 pKa = 5.91LSGGDD752 pKa = 3.66GNDD755 pKa = 3.22VFVSSPGNDD764 pKa = 3.32VIDD767 pKa = 4.78GGDD770 pKa = 3.63GSDD773 pKa = 3.26TLQLLGARR781 pKa = 11.84GDD783 pKa = 3.89YY784 pKa = 10.57TITASGAATLVEE796 pKa = 4.24GNAYY800 pKa = 10.12DD801 pKa = 4.12VGTLTLTSIEE811 pKa = 4.29NIRR814 pKa = 11.84FEE816 pKa = 5.03SDD818 pKa = 2.84DD819 pKa = 3.58ATYY822 pKa = 10.27TSQDD826 pKa = 2.86LFGYY830 pKa = 10.21YY831 pKa = 8.41GTAGDD836 pKa = 5.91DD837 pKa = 3.66VLLGNALGTNFYY849 pKa = 11.18ALAGDD854 pKa = 4.21DD855 pKa = 4.13QISPGIGNDD864 pKa = 3.38VVDD867 pKa = 4.72GGDD870 pKa = 3.75GEE872 pKa = 5.06DD873 pKa = 3.71EE874 pKa = 4.43VVLAASSANSSVSRR888 pKa = 11.84EE889 pKa = 3.79LDD891 pKa = 3.34GSISVYY897 pKa = 10.9DD898 pKa = 4.13PASGSYY904 pKa = 10.39DD905 pKa = 3.16ILWNVEE911 pKa = 3.8KK912 pKa = 10.59VTFLGDD918 pKa = 3.19STTIDD923 pKa = 3.66VGDD926 pKa = 4.73LPPLGTSGNDD936 pKa = 3.57TITGTARR943 pKa = 11.84GEE945 pKa = 4.08TLYY948 pKa = 11.21GQDD951 pKa = 4.92GDD953 pKa = 3.98DD954 pKa = 3.68QIYY957 pKa = 10.45GLGGNDD963 pKa = 4.02HH964 pKa = 7.11LNGGAGDD971 pKa = 4.34DD972 pKa = 4.39LLDD975 pKa = 4.76GGTGNNGLDD984 pKa = 3.78GGDD987 pKa = 4.22GNDD990 pKa = 3.8TLIGGAGQDD999 pKa = 2.99SMYY1002 pKa = 10.74GGEE1005 pKa = 4.61GSDD1008 pKa = 3.5TYY1010 pKa = 11.07IVGDD1014 pKa = 3.89AYY1016 pKa = 10.78GAFADD1021 pKa = 4.65TEE1023 pKa = 4.17VGDD1026 pKa = 3.83YY1027 pKa = 11.16GDD1029 pKa = 3.76YY1030 pKa = 11.5GNFVDD1035 pKa = 5.83HH1036 pKa = 7.36DD1037 pKa = 4.11TLALGDD1043 pKa = 3.75ATYY1046 pKa = 11.36SFAADD1051 pKa = 3.53GDD1053 pKa = 4.02RR1054 pKa = 11.84VIITNTATLHH1064 pKa = 5.36TVTLANQLSGYY1075 pKa = 8.57GVEE1078 pKa = 4.48SIEE1081 pKa = 4.21IGSNTVLDD1089 pKa = 4.3RR1090 pKa = 11.84DD1091 pKa = 5.18DD1092 pKa = 4.83ILDD1095 pKa = 5.33LIPVNVISGTSGDD1108 pKa = 3.99DD1109 pKa = 3.46QISGTNVNDD1118 pKa = 5.23DD1119 pKa = 3.27IAAGAGNDD1127 pKa = 3.93YY1128 pKa = 10.63IQEE1131 pKa = 4.2STGNDD1136 pKa = 2.99VYY1138 pKa = 10.94RR1139 pKa = 11.84WNVGDD1144 pKa = 3.85GDD1146 pKa = 4.59DD1147 pKa = 4.59YY1148 pKa = 11.62IAGGDD1153 pKa = 3.92SNDD1156 pKa = 3.88GYY1158 pKa = 11.52NSVVLGSGIATSDD1171 pKa = 3.43VTVSFTDD1178 pKa = 3.88FFEE1181 pKa = 4.5QSVLLSFAQGGSITLEE1197 pKa = 4.44NINGTDD1203 pKa = 3.43QNVDD1207 pKa = 3.18EE1208 pKa = 5.79VEE1210 pKa = 4.29FDD1212 pKa = 5.71DD1213 pKa = 3.83NTVWDD1218 pKa = 4.42RR1219 pKa = 11.84STLIGMAMDD1228 pKa = 3.69NSGGGGGGPLGLAANSKK1245 pKa = 8.86TGSFPAPHH1253 pKa = 6.25YY1254 pKa = 10.51RR1255 pKa = 11.84VDD1257 pKa = 3.4HH1258 pKa = 5.89NAVVSRR1264 pKa = 11.84TPVANSDD1271 pKa = 3.53VALDD1275 pKa = 4.02DD1276 pKa = 3.87FDD1278 pKa = 5.77YY1279 pKa = 10.85MGTARR1284 pKa = 11.84WAISKK1289 pKa = 10.6LSGYY1293 pKa = 10.98DD1294 pKa = 3.1EE1295 pKa = 4.12MVARR1299 pKa = 11.84KK1300 pKa = 9.57SRR1302 pKa = 11.84IAAGVSNRR1310 pKa = 11.84PFSGTDD1316 pKa = 2.81IAVSVAGNPSVNRR1329 pKa = 11.84SANLLAEE1336 pKa = 5.35AISTFDD1342 pKa = 3.22PTAAGGFSTSEE1353 pKa = 4.08DD1354 pKa = 3.09KK1355 pKa = 11.15GAPDD1359 pKa = 3.98NFWLFHH1365 pKa = 5.78QNGSLEE1371 pKa = 4.6SKK1373 pKa = 10.62SSLAMIAA1380 pKa = 3.71
MM1 pKa = 7.29TVPTEE6 pKa = 3.99PVTPEE11 pKa = 3.29MLLRR15 pKa = 11.84RR16 pKa = 11.84FLYY19 pKa = 10.23GVNEE23 pKa = 4.28TPDD26 pKa = 3.79LLTDD30 pKa = 4.15RR31 pKa = 11.84LRR33 pKa = 11.84DD34 pKa = 3.61LAPTASISIGEE45 pKa = 4.38INSDD49 pKa = 3.82TYY51 pKa = 10.38LASYY55 pKa = 9.75GRR57 pKa = 11.84YY58 pKa = 9.12INAAYY63 pKa = 9.93FPFVQDD69 pKa = 3.9FFTGAKK75 pKa = 7.66TLSVGTHH82 pKa = 5.52TVAQLQSTYY91 pKa = 10.62GASEE95 pKa = 3.91FLYY98 pKa = 10.38TLSNANTDD106 pKa = 3.3VGSSDD111 pKa = 3.32FFLRR115 pKa = 11.84AFMFGHH121 pKa = 7.43ADD123 pKa = 3.43FHH125 pKa = 7.01LSPDD129 pKa = 3.49TQFVVDD135 pKa = 4.07TNGDD139 pKa = 3.5LHH141 pKa = 8.22IEE143 pKa = 4.02NMRR146 pKa = 11.84YY147 pKa = 9.49IPDD150 pKa = 3.25VDD152 pKa = 4.52NFDD155 pKa = 4.31FDD157 pKa = 4.11STGPTAIYY165 pKa = 10.16DD166 pKa = 3.71AVFAKK171 pKa = 10.67PEE173 pKa = 4.17IDD175 pKa = 3.4PSSIGRR181 pKa = 11.84QITWTYY187 pKa = 10.02SDD189 pKa = 3.58PTIVHH194 pKa = 6.2NGYY197 pKa = 9.58YY198 pKa = 9.25AANYY202 pKa = 8.0AANVLNNYY210 pKa = 9.82NITSNSIYY218 pKa = 10.42AGAIILSQSFGYY230 pKa = 9.66FNQWKK235 pKa = 10.38SEE237 pKa = 3.61GVYY240 pKa = 10.95EE241 pKa = 4.43NFRR244 pKa = 11.84DD245 pKa = 3.89NKK247 pKa = 9.5TIIYY251 pKa = 9.81DD252 pKa = 3.7GLGTNVVVADD262 pKa = 3.49ITSLHH267 pKa = 6.03GSILIGGDD275 pKa = 3.78GNDD278 pKa = 3.63QLIGAHH284 pKa = 6.27YY285 pKa = 10.97DD286 pKa = 3.37DD287 pKa = 4.71VFYY290 pKa = 11.2GGDD293 pKa = 3.59GNDD296 pKa = 3.77TIKK299 pKa = 10.78PGLGSSEE306 pKa = 3.82LWGGDD311 pKa = 3.43EE312 pKa = 4.11EE313 pKa = 5.28SKK315 pKa = 10.99PEE317 pKa = 4.3YY318 pKa = 10.3EE319 pKa = 4.6DD320 pKa = 3.98TDD322 pKa = 3.73TADD325 pKa = 3.71YY326 pKa = 9.26TGHH329 pKa = 6.07SAIDD333 pKa = 3.27ITFDD337 pKa = 3.25NSGEE341 pKa = 4.07KK342 pKa = 10.53LSLKK346 pKa = 9.44VAHH349 pKa = 7.21DD350 pKa = 3.87GANDD354 pKa = 3.75TLHH357 pKa = 7.12SIEE360 pKa = 4.62VIKK363 pKa = 9.32GTPATDD369 pKa = 3.11HH370 pKa = 6.13VKK372 pKa = 10.46IIGSIPTATMLTIDD386 pKa = 3.69AAGGQGPNPLDD397 pKa = 3.94TINLAKK403 pKa = 10.13ADD405 pKa = 3.92NGIHH409 pKa = 6.42VSIANDD415 pKa = 3.56GTGYY419 pKa = 9.9IHH421 pKa = 8.11DD422 pKa = 4.49KK423 pKa = 8.03TTEE426 pKa = 3.81GDD428 pKa = 3.14ILLAGFNTQIIGSAFDD444 pKa = 5.37DD445 pKa = 4.18VFDD448 pKa = 5.91DD449 pKa = 4.21DD450 pKa = 5.35AVGAKK455 pKa = 10.11HH456 pKa = 6.95IDD458 pKa = 3.19GGYY461 pKa = 10.75GNDD464 pKa = 3.85TISITGASDD473 pKa = 4.05DD474 pKa = 3.92SLLLGGAGNDD484 pKa = 4.14TITGGDD490 pKa = 3.77GNDD493 pKa = 3.46VLVGGAYY500 pKa = 9.99AYY502 pKa = 9.97GAYY505 pKa = 10.24SDD507 pKa = 4.06TLNGGGGNDD516 pKa = 3.48MLIGSGANDD525 pKa = 3.44IFDD528 pKa = 4.29GGDD531 pKa = 3.28GYY533 pKa = 11.47DD534 pKa = 3.89YY535 pKa = 10.8IRR537 pKa = 11.84PTAIPGTTFGLATITGGAGDD557 pKa = 4.16DD558 pKa = 3.98VINLRR563 pKa = 11.84EE564 pKa = 4.44GGDD567 pKa = 3.76HH568 pKa = 6.08YY569 pKa = 11.72NGMYY573 pKa = 10.06HH574 pKa = 6.63YY575 pKa = 10.76GAAPTLNFGANDD587 pKa = 3.43GHH589 pKa = 6.44DD590 pKa = 3.83TVLTASGGDD599 pKa = 3.35VFTAYY604 pKa = 10.76DD605 pKa = 3.53NGVLNDD611 pKa = 4.15HH612 pKa = 6.54GADD615 pKa = 4.18GIVVKK620 pKa = 10.91LDD622 pKa = 3.11GLYY625 pKa = 10.85ASDD628 pKa = 5.37IDD630 pKa = 4.68IIWDD634 pKa = 3.42VTPQPEE640 pKa = 4.39PFFGLGFNGNGDD652 pKa = 3.7LAIVVKK658 pKa = 9.88STGASIFIEE667 pKa = 4.42DD668 pKa = 3.88VNGSATFGNQSSSSNHH684 pKa = 5.9FGVTILDD691 pKa = 3.84GAGNEE696 pKa = 3.43IWIPSFPVQYY706 pKa = 11.03GDD708 pKa = 3.26LSSYY712 pKa = 10.71RR713 pKa = 11.84GGEE716 pKa = 3.53AAYY719 pKa = 10.17FDD721 pKa = 4.23AAALSSSEE729 pKa = 4.17STGTSGDD736 pKa = 3.5DD737 pKa = 3.38VMSGGRR743 pKa = 11.84GDD745 pKa = 5.3DD746 pKa = 4.01DD747 pKa = 5.91LSGGDD752 pKa = 3.66GNDD755 pKa = 3.22VFVSSPGNDD764 pKa = 3.32VIDD767 pKa = 4.78GGDD770 pKa = 3.63GSDD773 pKa = 3.26TLQLLGARR781 pKa = 11.84GDD783 pKa = 3.89YY784 pKa = 10.57TITASGAATLVEE796 pKa = 4.24GNAYY800 pKa = 10.12DD801 pKa = 4.12VGTLTLTSIEE811 pKa = 4.29NIRR814 pKa = 11.84FEE816 pKa = 5.03SDD818 pKa = 2.84DD819 pKa = 3.58ATYY822 pKa = 10.27TSQDD826 pKa = 2.86LFGYY830 pKa = 10.21YY831 pKa = 8.41GTAGDD836 pKa = 5.91DD837 pKa = 3.66VLLGNALGTNFYY849 pKa = 11.18ALAGDD854 pKa = 4.21DD855 pKa = 4.13QISPGIGNDD864 pKa = 3.38VVDD867 pKa = 4.72GGDD870 pKa = 3.75GEE872 pKa = 5.06DD873 pKa = 3.71EE874 pKa = 4.43VVLAASSANSSVSRR888 pKa = 11.84EE889 pKa = 3.79LDD891 pKa = 3.34GSISVYY897 pKa = 10.9DD898 pKa = 4.13PASGSYY904 pKa = 10.39DD905 pKa = 3.16ILWNVEE911 pKa = 3.8KK912 pKa = 10.59VTFLGDD918 pKa = 3.19STTIDD923 pKa = 3.66VGDD926 pKa = 4.73LPPLGTSGNDD936 pKa = 3.57TITGTARR943 pKa = 11.84GEE945 pKa = 4.08TLYY948 pKa = 11.21GQDD951 pKa = 4.92GDD953 pKa = 3.98DD954 pKa = 3.68QIYY957 pKa = 10.45GLGGNDD963 pKa = 4.02HH964 pKa = 7.11LNGGAGDD971 pKa = 4.34DD972 pKa = 4.39LLDD975 pKa = 4.76GGTGNNGLDD984 pKa = 3.78GGDD987 pKa = 4.22GNDD990 pKa = 3.8TLIGGAGQDD999 pKa = 2.99SMYY1002 pKa = 10.74GGEE1005 pKa = 4.61GSDD1008 pKa = 3.5TYY1010 pKa = 11.07IVGDD1014 pKa = 3.89AYY1016 pKa = 10.78GAFADD1021 pKa = 4.65TEE1023 pKa = 4.17VGDD1026 pKa = 3.83YY1027 pKa = 11.16GDD1029 pKa = 3.76YY1030 pKa = 11.5GNFVDD1035 pKa = 5.83HH1036 pKa = 7.36DD1037 pKa = 4.11TLALGDD1043 pKa = 3.75ATYY1046 pKa = 11.36SFAADD1051 pKa = 3.53GDD1053 pKa = 4.02RR1054 pKa = 11.84VIITNTATLHH1064 pKa = 5.36TVTLANQLSGYY1075 pKa = 8.57GVEE1078 pKa = 4.48SIEE1081 pKa = 4.21IGSNTVLDD1089 pKa = 4.3RR1090 pKa = 11.84DD1091 pKa = 5.18DD1092 pKa = 4.83ILDD1095 pKa = 5.33LIPVNVISGTSGDD1108 pKa = 3.99DD1109 pKa = 3.46QISGTNVNDD1118 pKa = 5.23DD1119 pKa = 3.27IAAGAGNDD1127 pKa = 3.93YY1128 pKa = 10.63IQEE1131 pKa = 4.2STGNDD1136 pKa = 2.99VYY1138 pKa = 10.94RR1139 pKa = 11.84WNVGDD1144 pKa = 3.85GDD1146 pKa = 4.59DD1147 pKa = 4.59YY1148 pKa = 11.62IAGGDD1153 pKa = 3.92SNDD1156 pKa = 3.88GYY1158 pKa = 11.52NSVVLGSGIATSDD1171 pKa = 3.43VTVSFTDD1178 pKa = 3.88FFEE1181 pKa = 4.5QSVLLSFAQGGSITLEE1197 pKa = 4.44NINGTDD1203 pKa = 3.43QNVDD1207 pKa = 3.18EE1208 pKa = 5.79VEE1210 pKa = 4.29FDD1212 pKa = 5.71DD1213 pKa = 3.83NTVWDD1218 pKa = 4.42RR1219 pKa = 11.84STLIGMAMDD1228 pKa = 3.69NSGGGGGGPLGLAANSKK1245 pKa = 8.86TGSFPAPHH1253 pKa = 6.25YY1254 pKa = 10.51RR1255 pKa = 11.84VDD1257 pKa = 3.4HH1258 pKa = 5.89NAVVSRR1264 pKa = 11.84TPVANSDD1271 pKa = 3.53VALDD1275 pKa = 4.02DD1276 pKa = 3.87FDD1278 pKa = 5.77YY1279 pKa = 10.85MGTARR1284 pKa = 11.84WAISKK1289 pKa = 10.6LSGYY1293 pKa = 10.98DD1294 pKa = 3.1EE1295 pKa = 4.12MVARR1299 pKa = 11.84KK1300 pKa = 9.57SRR1302 pKa = 11.84IAAGVSNRR1310 pKa = 11.84PFSGTDD1316 pKa = 2.81IAVSVAGNPSVNRR1329 pKa = 11.84SANLLAEE1336 pKa = 5.35AISTFDD1342 pKa = 3.22PTAAGGFSTSEE1353 pKa = 4.08DD1354 pKa = 3.09KK1355 pKa = 11.15GAPDD1359 pKa = 3.98NFWLFHH1365 pKa = 5.78QNGSLEE1371 pKa = 4.6SKK1373 pKa = 10.62SSLAMIAA1380 pKa = 3.71
Molecular weight: 143.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8LD45|A0A5B8LD45_9SPHN DUF4405 domain-containing protein OS=Sphingomonas panacisoli OX=1813879 GN=FPZ24_00220 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.0KK41 pKa = 10.58LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84NRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.0KK41 pKa = 10.58LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
991115 |
29 |
1932 |
307.3 |
33.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.421 ± 0.059 | 0.729 ± 0.013 |
6.295 ± 0.034 | 4.876 ± 0.041 |
3.55 ± 0.027 | 8.932 ± 0.045 |
1.84 ± 0.023 | 5.107 ± 0.029 |
3.323 ± 0.035 | 9.511 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.022 | 2.664 ± 0.035 |
5.403 ± 0.035 | 2.926 ± 0.023 |
7.061 ± 0.044 | 5.103 ± 0.034 |
5.692 ± 0.036 | 7.367 ± 0.034 |
1.487 ± 0.021 | 2.279 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
