
Aquareovirus G (isolate American grass carp/USA/PB01-155/-) (AQRV-G)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Aquareovirus; Aquareovirus G; American grass carp reovirus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
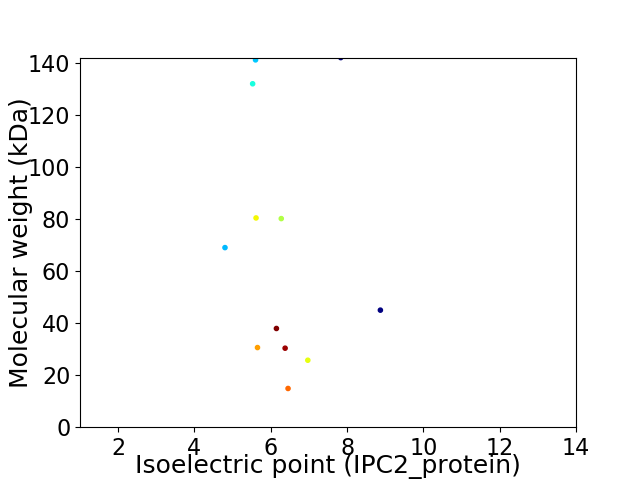
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|B2BNE5|VNS5_AQRVG Non-structural protein 5 OS=Aquareovirus G (isolate American grass carp/USA/PB01-155/-) OX=648234 GN=S7 PE=3 SV=1
MM1 pKa = 7.55GNVQTSTNVYY11 pKa = 10.5NIDD14 pKa = 3.52GNGNTFAPSSQMASTASPAIDD35 pKa = 4.29LKK37 pKa = 11.1PGVLNPTGKK46 pKa = 10.19LWQTMGTGAPSADD59 pKa = 3.4SLVLVVDD66 pKa = 3.85NKK68 pKa = 11.33GEE70 pKa = 3.97YY71 pKa = 9.6TYY73 pKa = 11.03LSEE76 pKa = 5.31NMRR79 pKa = 11.84EE80 pKa = 4.21TLNKK84 pKa = 10.32AVTDD88 pKa = 3.74VNMWQPLFQATKK100 pKa = 8.9SGCGPVVLANFTTISTGYY118 pKa = 10.29VGATADD124 pKa = 3.69DD125 pKa = 4.2AFSNGLVSNGPFLATMHH142 pKa = 6.35IMEE145 pKa = 4.61LQKK148 pKa = 10.04TIAARR153 pKa = 11.84MRR155 pKa = 11.84DD156 pKa = 3.39VAIWQKK162 pKa = 10.95HH163 pKa = 5.88LDD165 pKa = 3.52TAMTLMTPDD174 pKa = 3.43VSAGDD179 pKa = 3.97VTCKK183 pKa = 9.32WRR185 pKa = 11.84SLLEE189 pKa = 3.92FAQDD193 pKa = 4.42ILPLDD198 pKa = 4.03NLCRR202 pKa = 11.84SYY204 pKa = 11.29PNEE207 pKa = 4.2FYY209 pKa = 10.56TVAAQRR215 pKa = 11.84YY216 pKa = 5.68PAIRR220 pKa = 11.84PGQPDD225 pKa = 3.68TQVALPQPHH234 pKa = 7.45PLGEE238 pKa = 4.25VAGSFNAPTSEE249 pKa = 3.94VGSLVGAGAALSDD262 pKa = 5.11AISTLASKK270 pKa = 10.92DD271 pKa = 3.48LDD273 pKa = 3.73LVEE276 pKa = 5.92ADD278 pKa = 3.57TPLPVSVFTPSLAPRR293 pKa = 11.84TYY295 pKa = 10.02RR296 pKa = 11.84PAFIDD301 pKa = 3.88PQDD304 pKa = 4.07AAWIAQWNGDD314 pKa = 3.42ANIRR318 pKa = 11.84IITTYY323 pKa = 10.6QSTDD327 pKa = 3.17YY328 pKa = 10.24TVQLGPGPTRR338 pKa = 11.84VIDD341 pKa = 3.64MNAMIDD347 pKa = 3.49AKK349 pKa = 10.34LTLDD353 pKa = 3.39VSGTILPFQEE363 pKa = 4.58NNDD366 pKa = 3.65LSSAIPAFVLIQTKK380 pKa = 9.77VPLHH384 pKa = 6.41SVTQASDD391 pKa = 3.46VEE393 pKa = 4.8GITVVSAAEE402 pKa = 3.86SSAINLSVNVRR413 pKa = 11.84GDD415 pKa = 3.17PRR417 pKa = 11.84FDD419 pKa = 3.45MLHH422 pKa = 5.57LHH424 pKa = 7.01AMFEE428 pKa = 4.4RR429 pKa = 11.84EE430 pKa = 4.26TIAGIPYY437 pKa = 9.5IYY439 pKa = 10.63GIGTFLIPSITSSSSFCNPTLMDD462 pKa = 4.05GEE464 pKa = 4.58LTVTPLLLRR473 pKa = 11.84EE474 pKa = 4.17TTYY477 pKa = 11.06KK478 pKa = 10.62GAVVDD483 pKa = 4.02TVTPSEE489 pKa = 4.17VMANQTSEE497 pKa = 4.21EE498 pKa = 4.25VASALANDD506 pKa = 3.73AVLLVSGQLEE516 pKa = 3.94RR517 pKa = 11.84LATVVGDD524 pKa = 4.16VIPIASGEE532 pKa = 3.98DD533 pKa = 3.41DD534 pKa = 3.99AATSAIVGRR543 pKa = 11.84LAIEE547 pKa = 3.63ATMRR551 pKa = 11.84ARR553 pKa = 11.84HH554 pKa = 5.81GGDD557 pKa = 2.91TRR559 pKa = 11.84ALPNFGQLWKK569 pKa = 9.72RR570 pKa = 11.84AKK572 pKa = 10.02RR573 pKa = 11.84AASMFASNPALALQVGVPVLADD595 pKa = 3.52SGILSALTSGVSTAIRR611 pKa = 11.84TGSLGKK617 pKa = 10.43GVSDD621 pKa = 5.09ASSKK625 pKa = 11.0LNARR629 pKa = 11.84QSLTLARR636 pKa = 11.84KK637 pKa = 7.16TFFKK641 pKa = 10.52KK642 pKa = 10.69VEE644 pKa = 4.23EE645 pKa = 4.27LWPSQQ650 pKa = 3.26
MM1 pKa = 7.55GNVQTSTNVYY11 pKa = 10.5NIDD14 pKa = 3.52GNGNTFAPSSQMASTASPAIDD35 pKa = 4.29LKK37 pKa = 11.1PGVLNPTGKK46 pKa = 10.19LWQTMGTGAPSADD59 pKa = 3.4SLVLVVDD66 pKa = 3.85NKK68 pKa = 11.33GEE70 pKa = 3.97YY71 pKa = 9.6TYY73 pKa = 11.03LSEE76 pKa = 5.31NMRR79 pKa = 11.84EE80 pKa = 4.21TLNKK84 pKa = 10.32AVTDD88 pKa = 3.74VNMWQPLFQATKK100 pKa = 8.9SGCGPVVLANFTTISTGYY118 pKa = 10.29VGATADD124 pKa = 3.69DD125 pKa = 4.2AFSNGLVSNGPFLATMHH142 pKa = 6.35IMEE145 pKa = 4.61LQKK148 pKa = 10.04TIAARR153 pKa = 11.84MRR155 pKa = 11.84DD156 pKa = 3.39VAIWQKK162 pKa = 10.95HH163 pKa = 5.88LDD165 pKa = 3.52TAMTLMTPDD174 pKa = 3.43VSAGDD179 pKa = 3.97VTCKK183 pKa = 9.32WRR185 pKa = 11.84SLLEE189 pKa = 3.92FAQDD193 pKa = 4.42ILPLDD198 pKa = 4.03NLCRR202 pKa = 11.84SYY204 pKa = 11.29PNEE207 pKa = 4.2FYY209 pKa = 10.56TVAAQRR215 pKa = 11.84YY216 pKa = 5.68PAIRR220 pKa = 11.84PGQPDD225 pKa = 3.68TQVALPQPHH234 pKa = 7.45PLGEE238 pKa = 4.25VAGSFNAPTSEE249 pKa = 3.94VGSLVGAGAALSDD262 pKa = 5.11AISTLASKK270 pKa = 10.92DD271 pKa = 3.48LDD273 pKa = 3.73LVEE276 pKa = 5.92ADD278 pKa = 3.57TPLPVSVFTPSLAPRR293 pKa = 11.84TYY295 pKa = 10.02RR296 pKa = 11.84PAFIDD301 pKa = 3.88PQDD304 pKa = 4.07AAWIAQWNGDD314 pKa = 3.42ANIRR318 pKa = 11.84IITTYY323 pKa = 10.6QSTDD327 pKa = 3.17YY328 pKa = 10.24TVQLGPGPTRR338 pKa = 11.84VIDD341 pKa = 3.64MNAMIDD347 pKa = 3.49AKK349 pKa = 10.34LTLDD353 pKa = 3.39VSGTILPFQEE363 pKa = 4.58NNDD366 pKa = 3.65LSSAIPAFVLIQTKK380 pKa = 9.77VPLHH384 pKa = 6.41SVTQASDD391 pKa = 3.46VEE393 pKa = 4.8GITVVSAAEE402 pKa = 3.86SSAINLSVNVRR413 pKa = 11.84GDD415 pKa = 3.17PRR417 pKa = 11.84FDD419 pKa = 3.45MLHH422 pKa = 5.57LHH424 pKa = 7.01AMFEE428 pKa = 4.4RR429 pKa = 11.84EE430 pKa = 4.26TIAGIPYY437 pKa = 9.5IYY439 pKa = 10.63GIGTFLIPSITSSSSFCNPTLMDD462 pKa = 4.05GEE464 pKa = 4.58LTVTPLLLRR473 pKa = 11.84EE474 pKa = 4.17TTYY477 pKa = 11.06KK478 pKa = 10.62GAVVDD483 pKa = 4.02TVTPSEE489 pKa = 4.17VMANQTSEE497 pKa = 4.21EE498 pKa = 4.25VASALANDD506 pKa = 3.73AVLLVSGQLEE516 pKa = 3.94RR517 pKa = 11.84LATVVGDD524 pKa = 4.16VIPIASGEE532 pKa = 3.98DD533 pKa = 3.41DD534 pKa = 3.99AATSAIVGRR543 pKa = 11.84LAIEE547 pKa = 3.63ATMRR551 pKa = 11.84ARR553 pKa = 11.84HH554 pKa = 5.81GGDD557 pKa = 2.91TRR559 pKa = 11.84ALPNFGQLWKK569 pKa = 9.72RR570 pKa = 11.84AKK572 pKa = 10.02RR573 pKa = 11.84AASMFASNPALALQVGVPVLADD595 pKa = 3.52SGILSALTSGVSTAIRR611 pKa = 11.84TGSLGKK617 pKa = 10.43GVSDD621 pKa = 5.09ASSKK625 pKa = 11.0LNARR629 pKa = 11.84QSLTLARR636 pKa = 11.84KK637 pKa = 7.16TFFKK641 pKa = 10.52KK642 pKa = 10.69VEE644 pKa = 4.23EE645 pKa = 4.27LWPSQQ650 pKa = 3.26
Molecular weight: 68.99 kDa
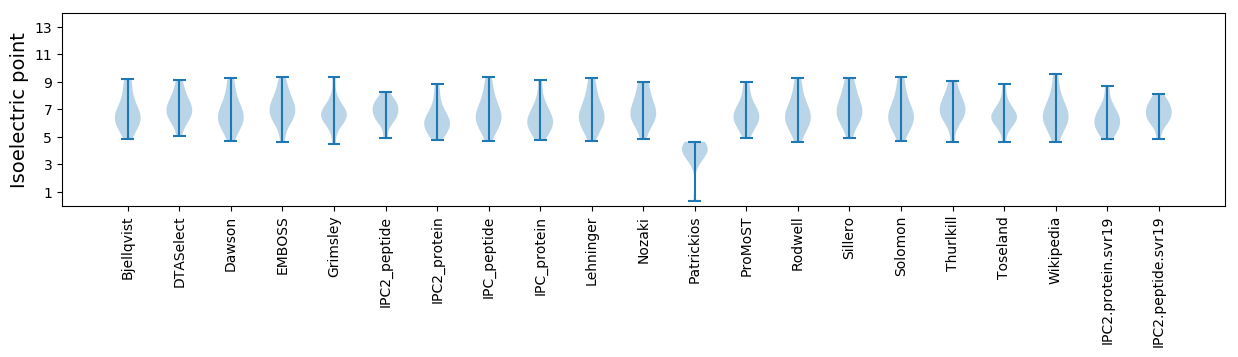
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B2BNE8|VNS2_AQRVG Non-structural protein 2 OS=Aquareovirus G (isolate American grass carp/USA/PB01-155/-) OX=648234 GN=S9 PE=3 SV=1
MM1 pKa = 7.71ALRR4 pKa = 11.84RR5 pKa = 11.84FLRR8 pKa = 11.84TSPITSTAQPAPLYY22 pKa = 10.43DD23 pKa = 3.64NQDD26 pKa = 2.96IQDD29 pKa = 3.33IVGAYY34 pKa = 8.68ARR36 pKa = 11.84PWQSRR41 pKa = 11.84FGDD44 pKa = 3.34ITITTTSAPVWSGRR58 pKa = 11.84YY59 pKa = 8.71PSVAARR65 pKa = 11.84NIIVNTILGAHH76 pKa = 6.8LNAFSGGVIAQYY88 pKa = 10.75RR89 pKa = 11.84GLTWRR94 pKa = 11.84DD95 pKa = 3.22NIMSSLAPPSQNPPPPAWVPAEE117 pKa = 3.85NVQLDD122 pKa = 3.8SDD124 pKa = 4.5NYY126 pKa = 8.95PQYY129 pKa = 11.75ALNLSKK135 pKa = 9.71MWSVNLDD142 pKa = 3.31VHH144 pKa = 8.13IMTMWALSDD153 pKa = 3.77YY154 pKa = 10.64GPLYY158 pKa = 10.01EE159 pKa = 4.96ISVPTAPMPAMTTAALMAYY178 pKa = 8.74IGCSITQLAMTAYY191 pKa = 9.79QYY193 pKa = 11.46AGQLPQTAAATMTTTLRR210 pKa = 11.84WLAAIWFGSLCGVVHH225 pKa = 7.32RR226 pKa = 11.84NHH228 pKa = 6.37TVNGFYY234 pKa = 10.55FDD236 pKa = 3.67FGKK239 pKa = 10.0PGFNPDD245 pKa = 3.12HH246 pKa = 7.19AVLKK250 pKa = 10.27WNDD253 pKa = 3.38GNRR256 pKa = 11.84AAPPAAARR264 pKa = 11.84FRR266 pKa = 11.84VYY268 pKa = 10.33RR269 pKa = 11.84VRR271 pKa = 11.84SPHH274 pKa = 4.7WQQMTSEE281 pKa = 4.32VAGAILAQSVTAVAGLTAMFNNRR304 pKa = 11.84GLPVWAQNIPHH315 pKa = 6.1FTGAAAGTRR324 pKa = 11.84VSRR327 pKa = 11.84TYY329 pKa = 11.41NPVTMAAARR338 pKa = 11.84HH339 pKa = 5.28QNWQAAGLITAVKK352 pKa = 9.91QAEE355 pKa = 4.36LDD357 pKa = 3.61QQYY360 pKa = 10.17TDD362 pKa = 3.45YY363 pKa = 11.24AQAIEE368 pKa = 4.17VHH370 pKa = 6.01LTAQLAANPVANGRR384 pKa = 11.84MPIQPFLPADD394 pKa = 4.26FAAAGGTNQVVADD407 pKa = 4.26ARR409 pKa = 11.84LMFPP413 pKa = 4.57
MM1 pKa = 7.71ALRR4 pKa = 11.84RR5 pKa = 11.84FLRR8 pKa = 11.84TSPITSTAQPAPLYY22 pKa = 10.43DD23 pKa = 3.64NQDD26 pKa = 2.96IQDD29 pKa = 3.33IVGAYY34 pKa = 8.68ARR36 pKa = 11.84PWQSRR41 pKa = 11.84FGDD44 pKa = 3.34ITITTTSAPVWSGRR58 pKa = 11.84YY59 pKa = 8.71PSVAARR65 pKa = 11.84NIIVNTILGAHH76 pKa = 6.8LNAFSGGVIAQYY88 pKa = 10.75RR89 pKa = 11.84GLTWRR94 pKa = 11.84DD95 pKa = 3.22NIMSSLAPPSQNPPPPAWVPAEE117 pKa = 3.85NVQLDD122 pKa = 3.8SDD124 pKa = 4.5NYY126 pKa = 8.95PQYY129 pKa = 11.75ALNLSKK135 pKa = 9.71MWSVNLDD142 pKa = 3.31VHH144 pKa = 8.13IMTMWALSDD153 pKa = 3.77YY154 pKa = 10.64GPLYY158 pKa = 10.01EE159 pKa = 4.96ISVPTAPMPAMTTAALMAYY178 pKa = 8.74IGCSITQLAMTAYY191 pKa = 9.79QYY193 pKa = 11.46AGQLPQTAAATMTTTLRR210 pKa = 11.84WLAAIWFGSLCGVVHH225 pKa = 7.32RR226 pKa = 11.84NHH228 pKa = 6.37TVNGFYY234 pKa = 10.55FDD236 pKa = 3.67FGKK239 pKa = 10.0PGFNPDD245 pKa = 3.12HH246 pKa = 7.19AVLKK250 pKa = 10.27WNDD253 pKa = 3.38GNRR256 pKa = 11.84AAPPAAARR264 pKa = 11.84FRR266 pKa = 11.84VYY268 pKa = 10.33RR269 pKa = 11.84VRR271 pKa = 11.84SPHH274 pKa = 4.7WQQMTSEE281 pKa = 4.32VAGAILAQSVTAVAGLTAMFNNRR304 pKa = 11.84GLPVWAQNIPHH315 pKa = 6.1FTGAAAGTRR324 pKa = 11.84VSRR327 pKa = 11.84TYY329 pKa = 11.41NPVTMAAARR338 pKa = 11.84HH339 pKa = 5.28QNWQAAGLITAVKK352 pKa = 9.91QAEE355 pKa = 4.36LDD357 pKa = 3.61QQYY360 pKa = 10.17TDD362 pKa = 3.45YY363 pKa = 11.24AQAIEE368 pKa = 4.17VHH370 pKa = 6.01LTAQLAANPVANGRR384 pKa = 11.84MPIQPFLPADD394 pKa = 4.26FAAAGGTNQVVADD407 pKa = 4.26ARR409 pKa = 11.84LMFPP413 pKa = 4.57
Molecular weight: 44.96 kDa
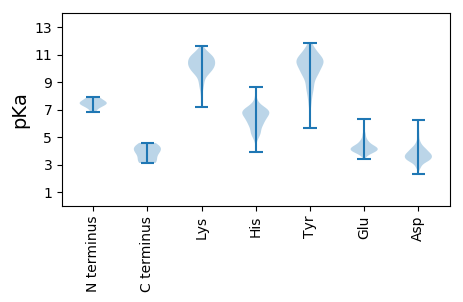
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7577 |
141 |
1298 |
631.4 |
69.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.703 ± 0.556 | 1.28 ± 0.175 |
5.147 ± 0.148 | 3.405 ± 0.396 |
3.062 ± 0.269 | 4.896 ± 0.335 |
2.732 ± 0.334 | 5.16 ± 0.222 |
1.98 ± 0.399 | 9.74 ± 0.382 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.679 ± 0.175 | 4.078 ± 0.434 |
7.958 ± 0.306 | 4.078 ± 0.154 |
5.728 ± 0.32 | 7.444 ± 0.553 |
8.42 ± 0.35 | 7.008 ± 0.314 |
1.412 ± 0.224 | 3.088 ± 0.39 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
