
Salinibacter ruber (strain DSM 13855 / M31)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidetes Order II. Incertae sedis; Rhodothermaceae; Salinibacter; Salinibacter ruber
Average proteome isoelectric point is 5.65
Get precalculated fractions of proteins
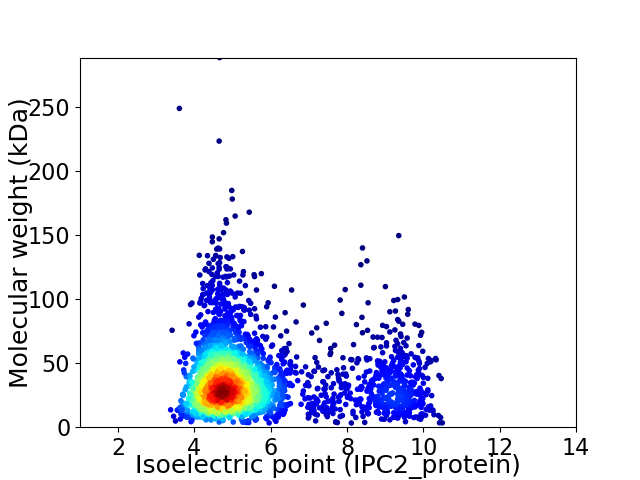
Virtual 2D-PAGE plot for 2812 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2S290|Q2S290_SALRD Uncharacterized protein OS=Salinibacter ruber (strain DSM 13855 / M31) OX=309807 GN=SRU_1570 PE=4 SV=1
MM1 pKa = 7.55SSDD4 pKa = 3.78PAPTDD9 pKa = 3.7TSPSWSDD16 pKa = 2.47RR17 pKa = 11.84WLNRR21 pKa = 11.84IEE23 pKa = 4.13RR24 pKa = 11.84TGNALPDD31 pKa = 3.81PVTLFFIFIAIVMVASWITHH51 pKa = 4.95TADD54 pKa = 2.85VSVVHH59 pKa = 6.94PGTDD63 pKa = 3.3EE64 pKa = 4.61TITADD69 pKa = 4.28NLFSDD74 pKa = 4.02EE75 pKa = 4.18NIRR78 pKa = 11.84RR79 pKa = 11.84LFTDD83 pKa = 3.18MAEE86 pKa = 4.36TFADD90 pKa = 5.38FPPLGLVLVVMLGIGVADD108 pKa = 3.47KK109 pKa = 10.88TGLISAALKK118 pKa = 10.74SFVASVPDD126 pKa = 3.93ALLTAALVFAGIMSSLAVDD145 pKa = 3.63AGYY148 pKa = 11.12VVVIPLGAVLFYY160 pKa = 11.12GVGRR164 pKa = 11.84HH165 pKa = 5.62PLAGLGAAFAGVSAGFSANLLLTSLDD191 pKa = 3.58PLLASFTEE199 pKa = 4.44PAAQLIAEE207 pKa = 4.95DD208 pKa = 3.96YY209 pKa = 10.35SVPVTANWYY218 pKa = 10.86LMIALTPVFVVLGAYY233 pKa = 8.07ITDD236 pKa = 4.7TIVEE240 pKa = 4.63PYY242 pKa = 9.9LGEE245 pKa = 4.14YY246 pKa = 9.11EE247 pKa = 4.16PPEE250 pKa = 4.64DD251 pKa = 4.67FDD253 pKa = 5.18EE254 pKa = 4.65EE255 pKa = 4.21DD256 pKa = 3.97AEE258 pKa = 4.63SSEE261 pKa = 4.15LTDD264 pKa = 3.25EE265 pKa = 4.24EE266 pKa = 4.62RR267 pKa = 11.84RR268 pKa = 11.84GLRR271 pKa = 11.84WAGGVTLASILGVVLLAVPEE291 pKa = 4.28GAPLAEE297 pKa = 4.46FEE299 pKa = 4.67ALIEE303 pKa = 4.51SIVALMVFLFFLPGLTYY320 pKa = 10.55GIVVGEE326 pKa = 4.54IEE328 pKa = 5.15DD329 pKa = 4.43DD330 pKa = 3.8SDD332 pKa = 3.99VADD335 pKa = 3.93MMADD339 pKa = 3.24SMADD343 pKa = 3.01MGAYY347 pKa = 9.0IVLAFAAAHH356 pKa = 6.34FIAMFEE362 pKa = 4.03WSNLGSIIAISGADD376 pKa = 3.41ALQSIGFTGLPLLFSFIFVSALINLFVGSASAKK409 pKa = 7.0WAIMAPVFVPMLMLAGDD426 pKa = 4.15PGYY429 pKa = 10.92SPEE432 pKa = 4.29TVQAAYY438 pKa = 10.51RR439 pKa = 11.84IGDD442 pKa = 3.78SFTNILTPLLPYY454 pKa = 9.98FPLVIIFAQRR464 pKa = 11.84YY465 pKa = 9.76DD466 pKa = 3.63EE467 pKa = 4.6DD468 pKa = 4.9AGIGSIIALMLPYY481 pKa = 10.28SVGFGVVSTIVFLVWVLLGLPLGPGAEE508 pKa = 4.26LYY510 pKa = 11.56YY511 pKa = 10.33MGG513 pKa = 5.26
MM1 pKa = 7.55SSDD4 pKa = 3.78PAPTDD9 pKa = 3.7TSPSWSDD16 pKa = 2.47RR17 pKa = 11.84WLNRR21 pKa = 11.84IEE23 pKa = 4.13RR24 pKa = 11.84TGNALPDD31 pKa = 3.81PVTLFFIFIAIVMVASWITHH51 pKa = 4.95TADD54 pKa = 2.85VSVVHH59 pKa = 6.94PGTDD63 pKa = 3.3EE64 pKa = 4.61TITADD69 pKa = 4.28NLFSDD74 pKa = 4.02EE75 pKa = 4.18NIRR78 pKa = 11.84RR79 pKa = 11.84LFTDD83 pKa = 3.18MAEE86 pKa = 4.36TFADD90 pKa = 5.38FPPLGLVLVVMLGIGVADD108 pKa = 3.47KK109 pKa = 10.88TGLISAALKK118 pKa = 10.74SFVASVPDD126 pKa = 3.93ALLTAALVFAGIMSSLAVDD145 pKa = 3.63AGYY148 pKa = 11.12VVVIPLGAVLFYY160 pKa = 11.12GVGRR164 pKa = 11.84HH165 pKa = 5.62PLAGLGAAFAGVSAGFSANLLLTSLDD191 pKa = 3.58PLLASFTEE199 pKa = 4.44PAAQLIAEE207 pKa = 4.95DD208 pKa = 3.96YY209 pKa = 10.35SVPVTANWYY218 pKa = 10.86LMIALTPVFVVLGAYY233 pKa = 8.07ITDD236 pKa = 4.7TIVEE240 pKa = 4.63PYY242 pKa = 9.9LGEE245 pKa = 4.14YY246 pKa = 9.11EE247 pKa = 4.16PPEE250 pKa = 4.64DD251 pKa = 4.67FDD253 pKa = 5.18EE254 pKa = 4.65EE255 pKa = 4.21DD256 pKa = 3.97AEE258 pKa = 4.63SSEE261 pKa = 4.15LTDD264 pKa = 3.25EE265 pKa = 4.24EE266 pKa = 4.62RR267 pKa = 11.84RR268 pKa = 11.84GLRR271 pKa = 11.84WAGGVTLASILGVVLLAVPEE291 pKa = 4.28GAPLAEE297 pKa = 4.46FEE299 pKa = 4.67ALIEE303 pKa = 4.51SIVALMVFLFFLPGLTYY320 pKa = 10.55GIVVGEE326 pKa = 4.54IEE328 pKa = 5.15DD329 pKa = 4.43DD330 pKa = 3.8SDD332 pKa = 3.99VADD335 pKa = 3.93MMADD339 pKa = 3.24SMADD343 pKa = 3.01MGAYY347 pKa = 9.0IVLAFAAAHH356 pKa = 6.34FIAMFEE362 pKa = 4.03WSNLGSIIAISGADD376 pKa = 3.41ALQSIGFTGLPLLFSFIFVSALINLFVGSASAKK409 pKa = 7.0WAIMAPVFVPMLMLAGDD426 pKa = 4.15PGYY429 pKa = 10.92SPEE432 pKa = 4.29TVQAAYY438 pKa = 10.51RR439 pKa = 11.84IGDD442 pKa = 3.78SFTNILTPLLPYY454 pKa = 9.98FPLVIIFAQRR464 pKa = 11.84YY465 pKa = 9.76DD466 pKa = 3.63EE467 pKa = 4.6DD468 pKa = 4.9AGIGSIIALMLPYY481 pKa = 10.28SVGFGVVSTIVFLVWVLLGLPLGPGAEE508 pKa = 4.26LYY510 pKa = 11.56YY511 pKa = 10.33MGG513 pKa = 5.26
Molecular weight: 54.51 kDa
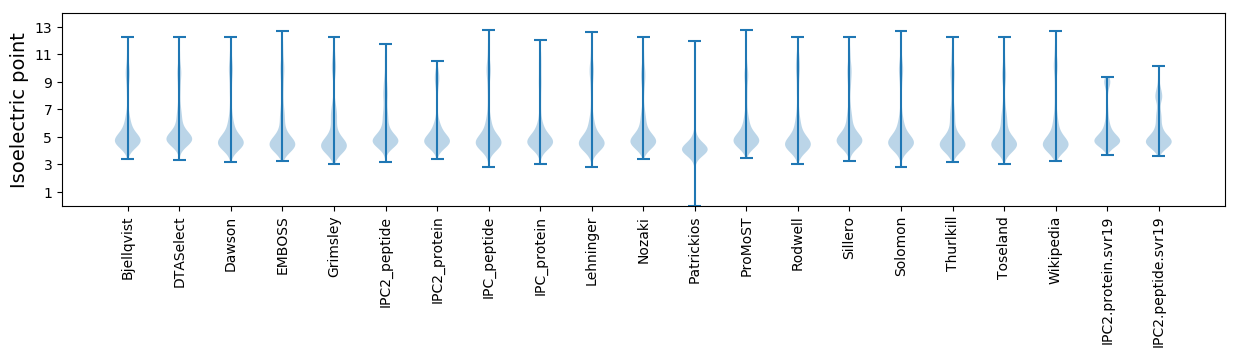
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2RYZ6|Q2RYZ6_SALRD Glycosyl hydrolase family 13 OS=Salinibacter ruber (strain DSM 13855 / M31) OX=309807 GN=SRU_2743 PE=4 SV=1
MM1 pKa = 7.91PPRR4 pKa = 11.84PTLRR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 4.18EE11 pKa = 4.19WTRR14 pKa = 11.84TRR16 pKa = 11.84NGEE19 pKa = 4.19VPRR22 pKa = 11.84HH23 pKa = 5.37FSLPSRR29 pKa = 11.84AGAAAHH35 pKa = 5.93EE36 pKa = 4.54ARR38 pKa = 11.84VSLAVAVEE46 pKa = 4.17EE47 pKa = 4.21AWARR51 pKa = 11.84CCRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PGRR59 pKa = 11.84EE60 pKa = 3.43RR61 pKa = 11.84LSPCHH66 pKa = 6.91DD67 pKa = 3.52GDD69 pKa = 4.49EE70 pKa = 4.67EE71 pKa = 4.67GTKK74 pKa = 9.58DD75 pKa = 3.21TAVRR79 pKa = 11.84ASARR83 pKa = 11.84GQPRR87 pKa = 11.84RR88 pKa = 11.84SCTPADD94 pKa = 3.75SRR96 pKa = 11.84ILAPTVARR104 pKa = 11.84VMSTSPDD111 pKa = 3.54DD112 pKa = 3.6NRR114 pKa = 11.84VWRR117 pKa = 11.84VFYY120 pKa = 10.54TRR122 pKa = 11.84ARR124 pKa = 11.84AEE126 pKa = 4.41KK127 pKa = 10.09KK128 pKa = 10.5CEE130 pKa = 3.72TRR132 pKa = 11.84LDD134 pKa = 3.5EE135 pKa = 4.36RR136 pKa = 11.84RR137 pKa = 11.84IDD139 pKa = 3.53VMVPKK144 pKa = 9.51KK145 pKa = 9.54TEE147 pKa = 3.47VRR149 pKa = 11.84QWSDD153 pKa = 2.6RR154 pKa = 11.84TKK156 pKa = 10.78EE157 pKa = 3.48ITEE160 pKa = 3.95PLFRR164 pKa = 11.84NYY166 pKa = 10.46LFARR170 pKa = 11.84VDD172 pKa = 3.29EE173 pKa = 4.35KK174 pKa = 11.53DD175 pKa = 3.07RR176 pKa = 11.84LRR178 pKa = 11.84VLRR181 pKa = 11.84TNGIVRR187 pKa = 11.84CVHH190 pKa = 6.66FDD192 pKa = 3.59GEE194 pKa = 4.31PARR197 pKa = 11.84LRR199 pKa = 11.84EE200 pKa = 4.08EE201 pKa = 4.2TVDD204 pKa = 3.66RR205 pKa = 11.84LQKK208 pKa = 10.36AQAVPEE214 pKa = 4.17RR215 pKa = 11.84LSTADD220 pKa = 3.46LRR222 pKa = 11.84PAVGEE227 pKa = 4.27TVTITGGPEE236 pKa = 3.57RR237 pKa = 11.84LQGLTGKK244 pKa = 9.52VLQHH248 pKa = 6.91RR249 pKa = 11.84GQTYY253 pKa = 10.59LLVQVRR259 pKa = 11.84AVRR262 pKa = 11.84QAVKK266 pKa = 10.45IEE268 pKa = 3.98VAADD272 pKa = 3.6WVQTTSNTT280 pKa = 3.44
MM1 pKa = 7.91PPRR4 pKa = 11.84PTLRR8 pKa = 11.84RR9 pKa = 11.84HH10 pKa = 4.18EE11 pKa = 4.19WTRR14 pKa = 11.84TRR16 pKa = 11.84NGEE19 pKa = 4.19VPRR22 pKa = 11.84HH23 pKa = 5.37FSLPSRR29 pKa = 11.84AGAAAHH35 pKa = 5.93EE36 pKa = 4.54ARR38 pKa = 11.84VSLAVAVEE46 pKa = 4.17EE47 pKa = 4.21AWARR51 pKa = 11.84CCRR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84PGRR59 pKa = 11.84EE60 pKa = 3.43RR61 pKa = 11.84LSPCHH66 pKa = 6.91DD67 pKa = 3.52GDD69 pKa = 4.49EE70 pKa = 4.67EE71 pKa = 4.67GTKK74 pKa = 9.58DD75 pKa = 3.21TAVRR79 pKa = 11.84ASARR83 pKa = 11.84GQPRR87 pKa = 11.84RR88 pKa = 11.84SCTPADD94 pKa = 3.75SRR96 pKa = 11.84ILAPTVARR104 pKa = 11.84VMSTSPDD111 pKa = 3.54DD112 pKa = 3.6NRR114 pKa = 11.84VWRR117 pKa = 11.84VFYY120 pKa = 10.54TRR122 pKa = 11.84ARR124 pKa = 11.84AEE126 pKa = 4.41KK127 pKa = 10.09KK128 pKa = 10.5CEE130 pKa = 3.72TRR132 pKa = 11.84LDD134 pKa = 3.5EE135 pKa = 4.36RR136 pKa = 11.84RR137 pKa = 11.84IDD139 pKa = 3.53VMVPKK144 pKa = 9.51KK145 pKa = 9.54TEE147 pKa = 3.47VRR149 pKa = 11.84QWSDD153 pKa = 2.6RR154 pKa = 11.84TKK156 pKa = 10.78EE157 pKa = 3.48ITEE160 pKa = 3.95PLFRR164 pKa = 11.84NYY166 pKa = 10.46LFARR170 pKa = 11.84VDD172 pKa = 3.29EE173 pKa = 4.35KK174 pKa = 11.53DD175 pKa = 3.07RR176 pKa = 11.84LRR178 pKa = 11.84VLRR181 pKa = 11.84TNGIVRR187 pKa = 11.84CVHH190 pKa = 6.66FDD192 pKa = 3.59GEE194 pKa = 4.31PARR197 pKa = 11.84LRR199 pKa = 11.84EE200 pKa = 4.08EE201 pKa = 4.2TVDD204 pKa = 3.66RR205 pKa = 11.84LQKK208 pKa = 10.36AQAVPEE214 pKa = 4.17RR215 pKa = 11.84LSTADD220 pKa = 3.46LRR222 pKa = 11.84PAVGEE227 pKa = 4.27TVTITGGPEE236 pKa = 3.57RR237 pKa = 11.84LQGLTGKK244 pKa = 9.52VLQHH248 pKa = 6.91RR249 pKa = 11.84GQTYY253 pKa = 10.59LLVQVRR259 pKa = 11.84AVRR262 pKa = 11.84QAVKK266 pKa = 10.45IEE268 pKa = 3.98VAADD272 pKa = 3.6WVQTTSNTT280 pKa = 3.44
Molecular weight: 31.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1007991 |
30 |
2597 |
358.5 |
39.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.703 ± 0.069 | 0.683 ± 0.012 |
7.015 ± 0.052 | 7.032 ± 0.07 |
3.394 ± 0.025 | 8.37 ± 0.046 |
2.227 ± 0.026 | 3.71 ± 0.03 |
2.105 ± 0.028 | 9.689 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.911 ± 0.018 | 2.451 ± 0.031 |
5.653 ± 0.038 | 3.549 ± 0.026 |
7.716 ± 0.06 | 5.833 ± 0.04 |
6.22 ± 0.03 | 7.872 ± 0.042 |
1.251 ± 0.018 | 2.617 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
