
Alistipes sp. An66
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; unclassified Alistipes
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
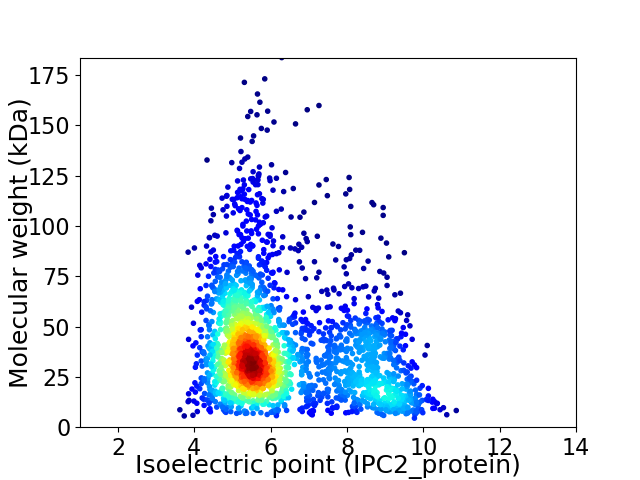
Virtual 2D-PAGE plot for 2381 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
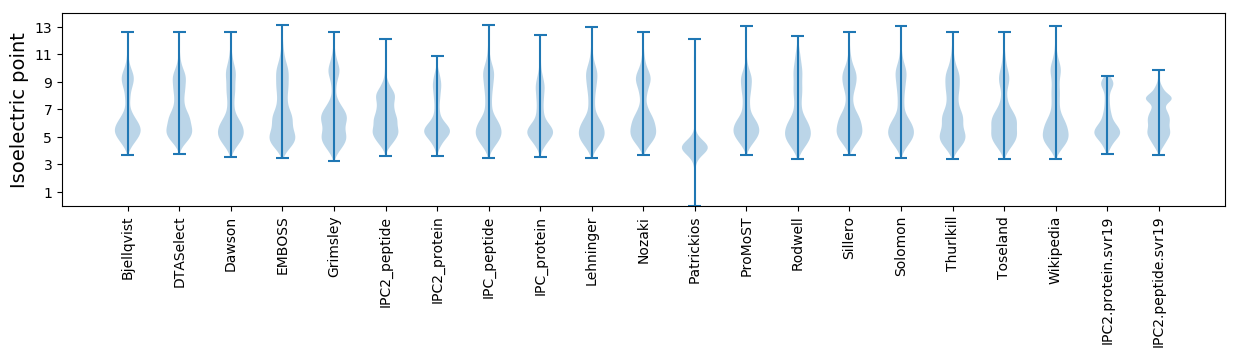
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3VGZ0|A0A1Y3VGZ0_9BACT V-type ATP synthase subunit D OS=Alistipes sp. An66 OX=1965650 GN=B5G16_02905 PE=3 SV=1
MM1 pKa = 7.61KK2 pKa = 10.47KK3 pKa = 10.33FLFLGWLLAAASLTACDD20 pKa = 4.72KK21 pKa = 11.44DD22 pKa = 3.83NEE24 pKa = 4.47QEE26 pKa = 4.96DD27 pKa = 5.4GPCPVTDD34 pKa = 3.45IEE36 pKa = 4.89LPQSSEE42 pKa = 3.73QTPIEE47 pKa = 4.49PGSTIEE53 pKa = 3.65IRR55 pKa = 11.84GNGFEE60 pKa = 4.89DD61 pKa = 4.53DD62 pKa = 3.95CEE64 pKa = 3.85IWLFGASQTAGVQAEE79 pKa = 4.17ILEE82 pKa = 4.47VSASAVRR89 pKa = 11.84FTAPNLCGTQRR100 pKa = 11.84IEE102 pKa = 3.61LRR104 pKa = 11.84QNGGVWDD111 pKa = 3.62IGTLVFPEE119 pKa = 4.43EE120 pKa = 3.95EE121 pKa = 4.06MLPIEE126 pKa = 4.16ILPRR130 pKa = 11.84RR131 pKa = 11.84ISHH134 pKa = 6.63IRR136 pKa = 11.84ITYY139 pKa = 9.66PDD141 pKa = 4.43DD142 pKa = 3.95LSFVQQFAYY151 pKa = 10.51DD152 pKa = 3.67EE153 pKa = 4.17QGRR156 pKa = 11.84IASITEE162 pKa = 3.49TDD164 pKa = 3.01QQYY167 pKa = 11.1AYY169 pKa = 9.74PGTTAQDD176 pKa = 3.28EE177 pKa = 4.73VTEE180 pKa = 4.09ITYY183 pKa = 10.52EE184 pKa = 3.78SDD186 pKa = 3.15RR187 pKa = 11.84IVITEE192 pKa = 3.82PSGAQSILTLADD204 pKa = 3.15GRR206 pKa = 11.84ATAITGVAFNDD217 pKa = 4.03NYY219 pKa = 10.53PDD221 pKa = 4.93DD222 pKa = 3.8YY223 pKa = 11.84AFTYY227 pKa = 10.09DD228 pKa = 3.35ANGYY232 pKa = 9.14IATSTWTQNPDD243 pKa = 2.63KK244 pKa = 10.18TYY246 pKa = 10.54SAKK249 pKa = 10.49SLYY252 pKa = 9.82TVVNGCMTQVRR263 pKa = 11.84EE264 pKa = 4.4AEE266 pKa = 5.27DD267 pKa = 3.19SGEE270 pKa = 4.09WYY272 pKa = 10.47GGNASYY278 pKa = 11.01TNNPEE283 pKa = 3.95RR284 pKa = 11.84MNNLNIDD291 pKa = 3.91LFGISDD297 pKa = 5.36FITTTDD303 pKa = 3.32LDD305 pKa = 4.0RR306 pKa = 11.84IYY308 pKa = 11.16LVGAGGNRR316 pKa = 11.84LKK318 pKa = 10.78KK319 pKa = 10.59LPASVDD325 pKa = 3.44YY326 pKa = 11.33GEE328 pKa = 5.25NGVDD332 pKa = 3.27TYY334 pKa = 11.02RR335 pKa = 11.84YY336 pKa = 7.75TMEE339 pKa = 4.49GEE341 pKa = 4.41YY342 pKa = 10.0ISKK345 pKa = 10.21IEE347 pKa = 3.89IDD349 pKa = 3.79EE350 pKa = 4.83DD351 pKa = 4.37GEE353 pKa = 4.19PLTILEE359 pKa = 4.6IFYY362 pKa = 11.07EE363 pKa = 4.41DD364 pKa = 3.2
MM1 pKa = 7.61KK2 pKa = 10.47KK3 pKa = 10.33FLFLGWLLAAASLTACDD20 pKa = 4.72KK21 pKa = 11.44DD22 pKa = 3.83NEE24 pKa = 4.47QEE26 pKa = 4.96DD27 pKa = 5.4GPCPVTDD34 pKa = 3.45IEE36 pKa = 4.89LPQSSEE42 pKa = 3.73QTPIEE47 pKa = 4.49PGSTIEE53 pKa = 3.65IRR55 pKa = 11.84GNGFEE60 pKa = 4.89DD61 pKa = 4.53DD62 pKa = 3.95CEE64 pKa = 3.85IWLFGASQTAGVQAEE79 pKa = 4.17ILEE82 pKa = 4.47VSASAVRR89 pKa = 11.84FTAPNLCGTQRR100 pKa = 11.84IEE102 pKa = 3.61LRR104 pKa = 11.84QNGGVWDD111 pKa = 3.62IGTLVFPEE119 pKa = 4.43EE120 pKa = 3.95EE121 pKa = 4.06MLPIEE126 pKa = 4.16ILPRR130 pKa = 11.84RR131 pKa = 11.84ISHH134 pKa = 6.63IRR136 pKa = 11.84ITYY139 pKa = 9.66PDD141 pKa = 4.43DD142 pKa = 3.95LSFVQQFAYY151 pKa = 10.51DD152 pKa = 3.67EE153 pKa = 4.17QGRR156 pKa = 11.84IASITEE162 pKa = 3.49TDD164 pKa = 3.01QQYY167 pKa = 11.1AYY169 pKa = 9.74PGTTAQDD176 pKa = 3.28EE177 pKa = 4.73VTEE180 pKa = 4.09ITYY183 pKa = 10.52EE184 pKa = 3.78SDD186 pKa = 3.15RR187 pKa = 11.84IVITEE192 pKa = 3.82PSGAQSILTLADD204 pKa = 3.15GRR206 pKa = 11.84ATAITGVAFNDD217 pKa = 4.03NYY219 pKa = 10.53PDD221 pKa = 4.93DD222 pKa = 3.8YY223 pKa = 11.84AFTYY227 pKa = 10.09DD228 pKa = 3.35ANGYY232 pKa = 9.14IATSTWTQNPDD243 pKa = 2.63KK244 pKa = 10.18TYY246 pKa = 10.54SAKK249 pKa = 10.49SLYY252 pKa = 9.82TVVNGCMTQVRR263 pKa = 11.84EE264 pKa = 4.4AEE266 pKa = 5.27DD267 pKa = 3.19SGEE270 pKa = 4.09WYY272 pKa = 10.47GGNASYY278 pKa = 11.01TNNPEE283 pKa = 3.95RR284 pKa = 11.84MNNLNIDD291 pKa = 3.91LFGISDD297 pKa = 5.36FITTTDD303 pKa = 3.32LDD305 pKa = 4.0RR306 pKa = 11.84IYY308 pKa = 11.16LVGAGGNRR316 pKa = 11.84LKK318 pKa = 10.78KK319 pKa = 10.59LPASVDD325 pKa = 3.44YY326 pKa = 11.33GEE328 pKa = 5.25NGVDD332 pKa = 3.27TYY334 pKa = 11.02RR335 pKa = 11.84YY336 pKa = 7.75TMEE339 pKa = 4.49GEE341 pKa = 4.41YY342 pKa = 10.0ISKK345 pKa = 10.21IEE347 pKa = 3.89IDD349 pKa = 3.79EE350 pKa = 4.83DD351 pKa = 4.37GEE353 pKa = 4.19PLTILEE359 pKa = 4.6IFYY362 pKa = 11.07EE363 pKa = 4.41DD364 pKa = 3.2
Molecular weight: 40.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3V9U3|A0A1Y3V9U3_9BACT NAD metabolism ATPase/kinase OS=Alistipes sp. An66 OX=1965650 GN=B5G16_11420 PE=4 SV=1
MM1 pKa = 7.27FKK3 pKa = 10.74KK4 pKa = 10.48RR5 pKa = 11.84MLIIPNLLMSLQRR18 pKa = 11.84SLKK21 pKa = 7.86TRR23 pKa = 11.84LWLVFVLMSFTRR35 pKa = 11.84SLILIIFLMLFMIWVIFLTSLLIFLLNWRR64 pKa = 11.84NIWW67 pKa = 3.32
MM1 pKa = 7.27FKK3 pKa = 10.74KK4 pKa = 10.48RR5 pKa = 11.84MLIIPNLLMSLQRR18 pKa = 11.84SLKK21 pKa = 7.86TRR23 pKa = 11.84LWLVFVLMSFTRR35 pKa = 11.84SLILIIFLMLFMIWVIFLTSLLIFLLNWRR64 pKa = 11.84NIWW67 pKa = 3.32
Molecular weight: 8.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
895525 |
38 |
1604 |
376.1 |
42.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.094 ± 0.055 | 1.279 ± 0.018 |
5.715 ± 0.034 | 6.686 ± 0.05 |
4.19 ± 0.028 | 7.61 ± 0.038 |
1.806 ± 0.022 | 5.722 ± 0.041 |
4.289 ± 0.039 | 9.218 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.446 ± 0.023 | 3.912 ± 0.036 |
4.263 ± 0.028 | 3.069 ± 0.023 |
6.841 ± 0.054 | 5.753 ± 0.041 |
5.842 ± 0.044 | 6.92 ± 0.038 |
1.388 ± 0.02 | 3.958 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
