
Novosphingobium sp. PC22D
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
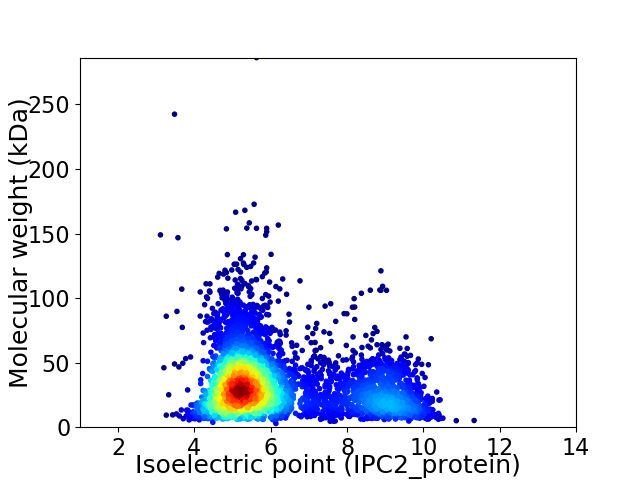
Virtual 2D-PAGE plot for 4593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A8HUX8|A0A2A8HUX8_9SPHN Zn-dependent hydrolase OS=Novosphingobium sp. PC22D OX=1962403 GN=B2G71_10410 PE=3 SV=1
MM1 pKa = 7.4KK2 pKa = 10.25NIHH5 pKa = 6.28GLLLVGCSALALAGCGPNEE24 pKa = 4.05IASPGTGGNITVNNPTPAPTPTPTPTPTPTSSLVTPAAGCPTIAASGGLGNEE76 pKa = 4.4GTISGPTGEE85 pKa = 4.58YY86 pKa = 9.52RR87 pKa = 11.84VCTLPTTFDD96 pKa = 3.35ASSTLPYY103 pKa = 10.13IEE105 pKa = 4.32GVLYY109 pKa = 10.84RR110 pKa = 11.84MNGRR114 pKa = 11.84VDD116 pKa = 3.37VGSDD120 pKa = 3.52GGPTPDD126 pKa = 5.27DD127 pKa = 3.81SDD129 pKa = 3.88GLSDD133 pKa = 3.75TNVTLKK139 pKa = 10.33IEE141 pKa = 4.41PGVIIYY147 pKa = 10.69SSGSSFLYY155 pKa = 10.3VNRR158 pKa = 11.84GNKK161 pKa = 8.7IDD163 pKa = 3.86AVGTKK168 pKa = 9.16MMPIVFTSRR177 pKa = 11.84DD178 pKa = 3.46NVQGLNNNNSSGQWGGVVLGGRR200 pKa = 11.84AQVTDD205 pKa = 3.85CDD207 pKa = 3.96APNAAEE213 pKa = 4.12GTIEE217 pKa = 5.07CEE219 pKa = 3.98RR220 pKa = 11.84QVEE223 pKa = 4.99GSATPAFFGGATNDD237 pKa = 3.79DD238 pKa = 3.98DD239 pKa = 5.09SGVMKK244 pKa = 10.15YY245 pKa = 8.5VQIRR249 pKa = 11.84YY250 pKa = 9.99SGFTLTGGSEE260 pKa = 4.19LQSLTTGGTGTGTEE274 pKa = 3.79FDD276 pKa = 4.7YY277 pKa = 11.2IQSFNSSDD285 pKa = 3.46DD286 pKa = 3.45GAEE289 pKa = 3.92FFGGYY294 pKa = 10.14VNMKK298 pKa = 9.69HH299 pKa = 6.37FIVVGAEE306 pKa = 3.95DD307 pKa = 4.94DD308 pKa = 5.35SIDD311 pKa = 3.71SDD313 pKa = 3.8TGVKK317 pKa = 10.66ANIQYY322 pKa = 10.75AIVVQDD328 pKa = 3.31SDD330 pKa = 3.82RR331 pKa = 11.84AHH333 pKa = 6.99DD334 pKa = 5.07AIIEE338 pKa = 4.18ADD340 pKa = 3.7SNDD343 pKa = 3.15NRR345 pKa = 11.84NAVPRR350 pKa = 11.84QDD352 pKa = 3.26TKK354 pKa = 10.66ISNFVFISPTAPADD368 pKa = 3.38AGAAVYY374 pKa = 9.86FRR376 pKa = 11.84GGTDD380 pKa = 3.27YY381 pKa = 11.87ALLNGIILSPNDD393 pKa = 2.8EE394 pKa = 4.95CIRR397 pKa = 11.84VRR399 pKa = 11.84HH400 pKa = 6.08AEE402 pKa = 4.17TVQASGDD409 pKa = 3.99DD410 pKa = 3.91EE411 pKa = 4.79NGPPVFEE418 pKa = 4.67SVQLSCSSAGQFVGSDD434 pKa = 3.36GPTADD439 pKa = 4.27DD440 pKa = 3.4VAGFFNAGSNNNATYY455 pKa = 10.34TSTLTNLFINGSNEE469 pKa = 3.83GGVTAFDD476 pKa = 3.98PTTLSSFFDD485 pKa = 3.33AVDD488 pKa = 3.51YY489 pKa = 11.16IGAVKK494 pKa = 10.32NAADD498 pKa = 3.15TWYY501 pKa = 9.54TGWTCNSDD509 pKa = 2.99IADD512 pKa = 4.17FGDD515 pKa = 3.44NTGACTSLPVYY526 pKa = 10.57
MM1 pKa = 7.4KK2 pKa = 10.25NIHH5 pKa = 6.28GLLLVGCSALALAGCGPNEE24 pKa = 4.05IASPGTGGNITVNNPTPAPTPTPTPTPTPTSSLVTPAAGCPTIAASGGLGNEE76 pKa = 4.4GTISGPTGEE85 pKa = 4.58YY86 pKa = 9.52RR87 pKa = 11.84VCTLPTTFDD96 pKa = 3.35ASSTLPYY103 pKa = 10.13IEE105 pKa = 4.32GVLYY109 pKa = 10.84RR110 pKa = 11.84MNGRR114 pKa = 11.84VDD116 pKa = 3.37VGSDD120 pKa = 3.52GGPTPDD126 pKa = 5.27DD127 pKa = 3.81SDD129 pKa = 3.88GLSDD133 pKa = 3.75TNVTLKK139 pKa = 10.33IEE141 pKa = 4.41PGVIIYY147 pKa = 10.69SSGSSFLYY155 pKa = 10.3VNRR158 pKa = 11.84GNKK161 pKa = 8.7IDD163 pKa = 3.86AVGTKK168 pKa = 9.16MMPIVFTSRR177 pKa = 11.84DD178 pKa = 3.46NVQGLNNNNSSGQWGGVVLGGRR200 pKa = 11.84AQVTDD205 pKa = 3.85CDD207 pKa = 3.96APNAAEE213 pKa = 4.12GTIEE217 pKa = 5.07CEE219 pKa = 3.98RR220 pKa = 11.84QVEE223 pKa = 4.99GSATPAFFGGATNDD237 pKa = 3.79DD238 pKa = 3.98DD239 pKa = 5.09SGVMKK244 pKa = 10.15YY245 pKa = 8.5VQIRR249 pKa = 11.84YY250 pKa = 9.99SGFTLTGGSEE260 pKa = 4.19LQSLTTGGTGTGTEE274 pKa = 3.79FDD276 pKa = 4.7YY277 pKa = 11.2IQSFNSSDD285 pKa = 3.46DD286 pKa = 3.45GAEE289 pKa = 3.92FFGGYY294 pKa = 10.14VNMKK298 pKa = 9.69HH299 pKa = 6.37FIVVGAEE306 pKa = 3.95DD307 pKa = 4.94DD308 pKa = 5.35SIDD311 pKa = 3.71SDD313 pKa = 3.8TGVKK317 pKa = 10.66ANIQYY322 pKa = 10.75AIVVQDD328 pKa = 3.31SDD330 pKa = 3.82RR331 pKa = 11.84AHH333 pKa = 6.99DD334 pKa = 5.07AIIEE338 pKa = 4.18ADD340 pKa = 3.7SNDD343 pKa = 3.15NRR345 pKa = 11.84NAVPRR350 pKa = 11.84QDD352 pKa = 3.26TKK354 pKa = 10.66ISNFVFISPTAPADD368 pKa = 3.38AGAAVYY374 pKa = 9.86FRR376 pKa = 11.84GGTDD380 pKa = 3.27YY381 pKa = 11.87ALLNGIILSPNDD393 pKa = 2.8EE394 pKa = 4.95CIRR397 pKa = 11.84VRR399 pKa = 11.84HH400 pKa = 6.08AEE402 pKa = 4.17TVQASGDD409 pKa = 3.99DD410 pKa = 3.91EE411 pKa = 4.79NGPPVFEE418 pKa = 4.67SVQLSCSSAGQFVGSDD434 pKa = 3.36GPTADD439 pKa = 4.27DD440 pKa = 3.4VAGFFNAGSNNNATYY455 pKa = 10.34TSTLTNLFINGSNEE469 pKa = 3.83GGVTAFDD476 pKa = 3.98PTTLSSFFDD485 pKa = 3.33AVDD488 pKa = 3.51YY489 pKa = 11.16IGAVKK494 pKa = 10.32NAADD498 pKa = 3.15TWYY501 pKa = 9.54TGWTCNSDD509 pKa = 2.99IADD512 pKa = 4.17FGDD515 pKa = 3.44NTGACTSLPVYY526 pKa = 10.57
Molecular weight: 54.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A8I046|A0A2A8I046_9SPHN HTH luxR-type domain-containing protein OS=Novosphingobium sp. PC22D OX=1962403 GN=B2G71_00260 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.57GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.1ATPGGRR28 pKa = 11.84KK29 pKa = 8.0VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84KK13 pKa = 9.21RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.57GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.1ATPGGRR28 pKa = 11.84KK29 pKa = 8.0VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1478106 |
25 |
2582 |
321.8 |
34.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.086 ± 0.053 | 0.901 ± 0.013 |
6.062 ± 0.033 | 6.132 ± 0.031 |
3.618 ± 0.021 | 8.821 ± 0.034 |
1.996 ± 0.018 | 4.852 ± 0.023 |
2.89 ± 0.031 | 9.849 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.45 ± 0.022 | 2.402 ± 0.023 |
5.241 ± 0.024 | 2.933 ± 0.018 |
7.5 ± 0.044 | 5.35 ± 0.031 |
5.064 ± 0.029 | 7.135 ± 0.027 |
1.47 ± 0.015 | 2.248 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
