
Solimonas terrae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Nevskiales; Sinobacteraceae; Solimonas
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
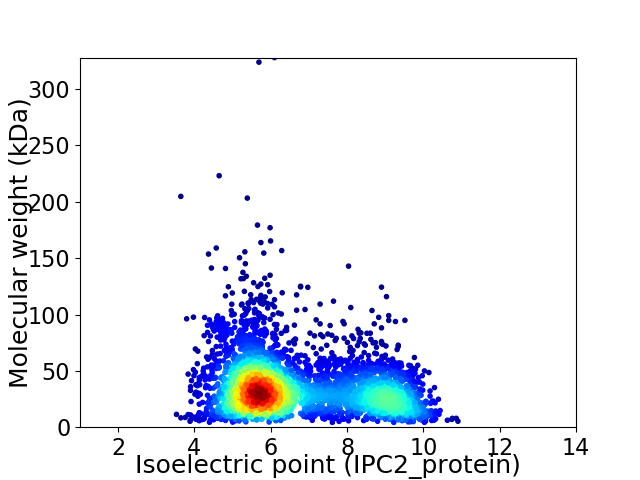
Virtual 2D-PAGE plot for 3931 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M2BPU4|A0A6M2BPU4_9GAMM Uncharacterized protein (Fragment) OS=Solimonas terrae OX=1396819 GN=G7Y85_06010 PE=4 SV=1
MM1 pKa = 7.41KK2 pKa = 9.79PYY4 pKa = 10.2RR5 pKa = 11.84LKK7 pKa = 10.88LHH9 pKa = 7.42PITAALRR16 pKa = 11.84FGARR20 pKa = 11.84GAVGSLLPGLALANPSGGVVVAGQAGINSEE50 pKa = 4.23GTQTTITQSSQSAVVNWQSFSVGSNEE76 pKa = 3.24YY77 pKa = 10.87VVFHH81 pKa = 6.05QPNASAAILNRR92 pKa = 11.84VVGGSPSDD100 pKa = 3.28ILGSISANGRR110 pKa = 11.84VFIINPHH117 pKa = 4.7GVMFGQNSRR126 pKa = 11.84IDD128 pKa = 3.73VGSLVATTMDD138 pKa = 3.12ISNADD143 pKa = 3.71FQAGRR148 pKa = 11.84YY149 pKa = 7.16QFVDD153 pKa = 3.99GSGDD157 pKa = 3.61GKK159 pKa = 11.02VDD161 pKa = 3.29NAGVITASNGGFVVLAADD179 pKa = 3.48QVGNSGLIQAQLGDD193 pKa = 3.43IALASGSALTLNLDD207 pKa = 3.67SEE209 pKa = 4.79GLVNFSIDD217 pKa = 3.22GAALSDD223 pKa = 3.48AAGVRR228 pKa = 11.84NLGTLAANGGRR239 pKa = 11.84VYY241 pKa = 10.38MSADD245 pKa = 3.27VARR248 pKa = 11.84QLIGTVVNNQGRR260 pKa = 11.84VSAQSIEE267 pKa = 4.11EE268 pKa = 3.88HH269 pKa = 7.04DD270 pKa = 4.01GQIVLSASGGDD281 pKa = 3.25IAQSGTVDD289 pKa = 3.15ASGDD293 pKa = 3.54GSGDD297 pKa = 3.41GGSVQITGTGDD308 pKa = 2.72IALAAGSLTQAGSASAHH325 pKa = 5.83GGSVAVAGDD334 pKa = 3.54GDD336 pKa = 3.73VDD338 pKa = 4.37FDD340 pKa = 3.58AGARR344 pKa = 11.84LDD346 pKa = 3.56VAGAQGGSVEE356 pKa = 4.5LSGHH360 pKa = 5.59RR361 pKa = 11.84NLRR364 pKa = 11.84IRR366 pKa = 11.84GDD368 pKa = 3.71LEE370 pKa = 4.41LGNGGSLLIDD380 pKa = 3.6PTTVTIAQGQGSTDD394 pKa = 3.38NGSDD398 pKa = 2.97ATVFEE403 pKa = 4.3QTLEE407 pKa = 3.99GLLAQGASVYY417 pKa = 10.1IAADD421 pKa = 3.56QAIHH425 pKa = 6.24VADD428 pKa = 5.05LSANGGDD435 pKa = 4.04GILDD439 pKa = 3.71GRR441 pKa = 11.84AGGYY445 pKa = 9.86GGSLTLGIGTQGYY458 pKa = 9.81GGAFGTGTGFSRR470 pKa = 11.84GDD472 pKa = 3.55GVSGGGIFFDD482 pKa = 3.7NTANTIAVDD491 pKa = 3.45GDD493 pKa = 3.77LAILGGYY500 pKa = 6.64TAGEE504 pKa = 4.07LTLGNLIGNSIDD516 pKa = 5.1LEE518 pKa = 4.28AADD521 pKa = 5.59AIHH524 pKa = 6.94ALGSITANNNGQIYY538 pKa = 9.91IDD540 pKa = 3.71STGGDD545 pKa = 3.7VTTHH549 pKa = 6.72ALNGGDD555 pKa = 3.03ITVSAEE561 pKa = 3.96TGALAALGDD570 pKa = 3.82ITISSGTGTVDD581 pKa = 3.74LRR583 pKa = 11.84AGHH586 pKa = 7.22DD587 pKa = 3.65LTVGAITIDD596 pKa = 3.48AGSAPQSYY604 pKa = 10.06GGSIDD609 pKa = 3.48YY610 pKa = 11.16GSIRR614 pKa = 11.84LSADD618 pKa = 3.49DD619 pKa = 4.79GDD621 pKa = 4.54LSVDD625 pKa = 3.3GATLVGDD632 pKa = 4.4LEE634 pKa = 5.22ASLSVAYY641 pKa = 10.27GGTPGLLSITDD652 pKa = 3.8STIEE656 pKa = 4.77GYY658 pKa = 7.79TALYY662 pKa = 9.73AGNGAGITLGGVTINDD678 pKa = 3.36NGLYY682 pKa = 10.6ASVSDD687 pKa = 4.45GGALSADD694 pKa = 3.51NLTASDD700 pKa = 4.43IYY702 pKa = 10.41LTSNSSIDD710 pKa = 3.6TSQGASQAAMQLGNLTATDD729 pKa = 3.35GGLYY733 pKa = 9.51IDD735 pKa = 4.83AGGDD739 pKa = 2.88ITTLHH744 pKa = 6.65NGTALIQGSEE754 pKa = 4.03NVEE757 pKa = 3.93IYY759 pKa = 10.66AGEE762 pKa = 4.44SYY764 pKa = 11.01DD765 pKa = 4.09SEE767 pKa = 4.51TGTTVGGNITLDD779 pKa = 3.74SVHH782 pKa = 6.87AGYY785 pKa = 10.76GIDD788 pKa = 3.99ISNDD792 pKa = 2.95SASYY796 pKa = 9.89YY797 pKa = 10.85GGAFTVGSHH806 pKa = 7.67DD807 pKa = 3.13ITFAHH812 pKa = 6.57LTADD816 pKa = 3.32AGSIYY821 pKa = 9.37VTAEE825 pKa = 3.9RR826 pKa = 11.84NVLATDD832 pKa = 4.12PASSTILATDD842 pKa = 3.6VAYY845 pKa = 10.79LSGEE849 pKa = 3.93NLTIGDD855 pKa = 3.83VSADD859 pKa = 3.65SVDD862 pKa = 3.48LNAYY866 pKa = 9.76GGTINTGAITAFAGNSYY883 pKa = 10.96SYY885 pKa = 11.48GDD887 pKa = 4.54DD888 pKa = 3.16IFFGLHH894 pKa = 6.65DD895 pKa = 4.94LGEE898 pKa = 4.08AHH900 pKa = 6.79GAIGTTGPIRR910 pKa = 11.84VTGGAGSVAIFADD923 pKa = 2.91SGYY926 pKa = 11.05AVSGLNIDD934 pKa = 4.3AGIDD938 pKa = 3.63PVDD941 pKa = 3.63GTRR944 pKa = 11.84GSIEE948 pKa = 4.05FFSSLGSLSVTDD960 pKa = 3.81STLHH964 pKa = 6.42GDD966 pKa = 3.51VTALLLGYY974 pKa = 9.94SDD976 pKa = 3.5QSYY979 pKa = 11.14GGNQLGTLAVGVPTVGDD996 pKa = 3.56DD997 pKa = 3.21KK998 pKa = 11.74LYY1000 pKa = 10.39IDD1002 pKa = 5.11GVAIDD1007 pKa = 4.7GDD1009 pKa = 4.1LVAYY1013 pKa = 9.22TDD1015 pKa = 3.77GGQAIDD1021 pKa = 4.11IGTLTATGEE1030 pKa = 4.2AVIGEE1035 pKa = 4.48YY1036 pKa = 10.77SSVPASSITALDD1048 pKa = 3.27ITAGGIEE1055 pKa = 4.37LGRR1058 pKa = 11.84DD1059 pKa = 3.36FAYY1062 pKa = 10.51GGTVGTDD1069 pKa = 2.85ILVGSLHH1076 pKa = 6.08ATDD1079 pKa = 4.54GDD1081 pKa = 4.11VEE1083 pKa = 4.21ILAGRR1088 pKa = 11.84NIGIISGSTGTVTADD1103 pKa = 2.93QGGVYY1108 pKa = 10.29LSAGTAQIDD1117 pKa = 3.68TGNGYY1122 pKa = 10.08AAGGGDD1128 pKa = 4.05IEE1130 pKa = 5.8IGDD1133 pKa = 3.73LQARR1137 pKa = 11.84QVAIDD1142 pKa = 4.47LYY1144 pKa = 11.01GGQLDD1149 pKa = 3.85TGAITVGGGSDD1160 pKa = 3.74YY1161 pKa = 11.16YY1162 pKa = 10.98DD1163 pKa = 3.4VYY1165 pKa = 11.28FGFHH1169 pKa = 7.01KK1170 pKa = 10.89DD1171 pKa = 3.12SDD1173 pKa = 4.67SIAHH1177 pKa = 6.95IGTIEE1182 pKa = 4.39SISVTDD1188 pKa = 3.8HH1189 pKa = 6.74AGHH1192 pKa = 6.45VYY1194 pKa = 10.14IGSDD1198 pKa = 3.4GDD1200 pKa = 4.15LDD1202 pKa = 3.48IASIGRR1208 pKa = 11.84IDD1210 pKa = 4.17SGSGTDD1216 pKa = 4.03GYY1218 pKa = 11.63GGTTLGSVSIGSEE1231 pKa = 4.0GGDD1234 pKa = 3.17LRR1236 pKa = 11.84VNALDD1241 pKa = 3.93VIGDD1245 pKa = 3.91LSIHH1249 pKa = 4.99MTNVIAHH1256 pKa = 7.3DD1257 pKa = 4.14GNGIALTASGSTSIFANMAAQDD1279 pKa = 3.51VHH1281 pKa = 8.26IDD1283 pKa = 3.19AGRR1286 pKa = 11.84DD1287 pKa = 3.26ISVDD1291 pKa = 4.13DD1292 pKa = 4.43IDD1294 pKa = 5.03AGSDD1298 pKa = 3.48GSVALLAHH1306 pKa = 7.2DD1307 pKa = 4.06GDD1309 pKa = 4.69LYY1311 pKa = 11.44RR1312 pKa = 11.84NGTISTADD1320 pKa = 3.76LSLQANSLSLDD1331 pKa = 4.17PIVLSGSLSLTSNTGGIEE1349 pKa = 3.98LGGAQASSIDD1359 pKa = 3.51LHH1361 pKa = 6.32AANGIDD1367 pKa = 5.14SGDD1370 pKa = 3.85LNAGSSGSIVLVADD1384 pKa = 3.97SGAITTGALTTGNLSVTATSLDD1406 pKa = 3.72FGDD1409 pKa = 3.57VDD1411 pKa = 3.63IAGDD1415 pKa = 4.06LSLTASGGDD1424 pKa = 3.34LSVGNLTAGSVSLSATDD1441 pKa = 3.6ALSVGGQIGSSGATSLNGASIALADD1466 pKa = 4.14QITGSSITIDD1476 pKa = 3.2GGALSGILDD1485 pKa = 4.23LVAGAGGIALDD1496 pKa = 4.06NSSGTVDD1503 pKa = 3.42SASLSSTGAVGIVGPLTATGAIAIDD1528 pKa = 3.95GGSIALADD1536 pKa = 3.76QLSASSITIDD1546 pKa = 3.95GGALSGKK1553 pKa = 9.98LDD1555 pKa = 3.76LQAGPGGIALHH1566 pKa = 6.2NSGSNVTSAKK1576 pKa = 10.06LDD1578 pKa = 3.62SGSKK1582 pKa = 9.93ISIAASLTASGNVTLNAAGSADD1604 pKa = 3.59FANITAHH1611 pKa = 6.89NITIDD1616 pKa = 3.53AGSDD1620 pKa = 3.19STLTHH1625 pKa = 6.48GSVLTAASGKK1635 pKa = 8.98VHH1637 pKa = 7.08LGGPSFSGSSLTVASASLDD1656 pKa = 3.85LASDD1660 pKa = 4.61LYY1662 pKa = 11.7VDD1664 pKa = 4.85ALDD1667 pKa = 4.43IDD1669 pKa = 4.05VSGANGLSLDD1679 pKa = 3.76NVLLSGTSIDD1689 pKa = 4.06LGATHH1694 pKa = 7.86GITLDD1699 pKa = 4.18DD1700 pKa = 3.88LTIDD1704 pKa = 4.0ASQIGLTAGDD1714 pKa = 4.17TLTATDD1720 pKa = 3.78TVLAGNLTATAGGTAQIDD1738 pKa = 3.46NATWQGGTASLEE1750 pKa = 4.09AASLDD1755 pKa = 3.44IGNSQLAYY1763 pKa = 7.84TQSASFITSDD1773 pKa = 3.98GSLNLIDD1780 pKa = 5.07SSFGPISTVQPLAATQSPTVGSGNLSLQSAGDD1812 pKa = 3.67IVLGGSDD1819 pKa = 3.1IRR1821 pKa = 11.84SGRR1824 pKa = 11.84LTLTAAGGILDD1835 pKa = 5.31DD1836 pKa = 5.11GSAANIDD1843 pKa = 3.66VDD1845 pKa = 4.47ALRR1848 pKa = 11.84ANAGTAIDD1856 pKa = 4.19FARR1859 pKa = 11.84SSLIIGTGSSGVAGDD1874 pKa = 3.99SGLLQLLAAQAPQLLPTATAPNGSLHH1900 pKa = 6.09AQSIALGTLAMSGDD1914 pKa = 4.25YY1915 pKa = 11.1LSLTTNALTIGHH1927 pKa = 6.35VSAAPAHH1934 pKa = 6.54LLVQIQPLAATTGIEE1949 pKa = 4.08LSQPTTVAARR1959 pKa = 11.84ALSSPAALNFSDD1971 pKa = 4.05AAAFAPYY1978 pKa = 9.79DD1979 pKa = 3.46ASLAFGGSNYY1989 pKa = 10.7AGDD1992 pKa = 4.0INVDD1996 pKa = 3.46SGVDD2000 pKa = 3.41VANASTNFLFMTSGHH2015 pKa = 5.56VNGHH2019 pKa = 6.39RR2020 pKa = 11.84NLATNGQVIVLSGIVTTEE2038 pKa = 3.7NLSDD2042 pKa = 4.23FEE2044 pKa = 4.82LPLIGEE2050 pKa = 4.24IQSGGLGDD2058 pKa = 4.63TPPLDD2063 pKa = 5.06DD2064 pKa = 5.29NPAAHH2069 pKa = 7.46PDD2071 pKa = 3.81DD2072 pKa = 4.18TSSLITIEE2080 pKa = 4.66DD2081 pKa = 3.8APGDD2085 pKa = 4.03TQCRR2089 pKa = 3.74
MM1 pKa = 7.41KK2 pKa = 9.79PYY4 pKa = 10.2RR5 pKa = 11.84LKK7 pKa = 10.88LHH9 pKa = 7.42PITAALRR16 pKa = 11.84FGARR20 pKa = 11.84GAVGSLLPGLALANPSGGVVVAGQAGINSEE50 pKa = 4.23GTQTTITQSSQSAVVNWQSFSVGSNEE76 pKa = 3.24YY77 pKa = 10.87VVFHH81 pKa = 6.05QPNASAAILNRR92 pKa = 11.84VVGGSPSDD100 pKa = 3.28ILGSISANGRR110 pKa = 11.84VFIINPHH117 pKa = 4.7GVMFGQNSRR126 pKa = 11.84IDD128 pKa = 3.73VGSLVATTMDD138 pKa = 3.12ISNADD143 pKa = 3.71FQAGRR148 pKa = 11.84YY149 pKa = 7.16QFVDD153 pKa = 3.99GSGDD157 pKa = 3.61GKK159 pKa = 11.02VDD161 pKa = 3.29NAGVITASNGGFVVLAADD179 pKa = 3.48QVGNSGLIQAQLGDD193 pKa = 3.43IALASGSALTLNLDD207 pKa = 3.67SEE209 pKa = 4.79GLVNFSIDD217 pKa = 3.22GAALSDD223 pKa = 3.48AAGVRR228 pKa = 11.84NLGTLAANGGRR239 pKa = 11.84VYY241 pKa = 10.38MSADD245 pKa = 3.27VARR248 pKa = 11.84QLIGTVVNNQGRR260 pKa = 11.84VSAQSIEE267 pKa = 4.11EE268 pKa = 3.88HH269 pKa = 7.04DD270 pKa = 4.01GQIVLSASGGDD281 pKa = 3.25IAQSGTVDD289 pKa = 3.15ASGDD293 pKa = 3.54GSGDD297 pKa = 3.41GGSVQITGTGDD308 pKa = 2.72IALAAGSLTQAGSASAHH325 pKa = 5.83GGSVAVAGDD334 pKa = 3.54GDD336 pKa = 3.73VDD338 pKa = 4.37FDD340 pKa = 3.58AGARR344 pKa = 11.84LDD346 pKa = 3.56VAGAQGGSVEE356 pKa = 4.5LSGHH360 pKa = 5.59RR361 pKa = 11.84NLRR364 pKa = 11.84IRR366 pKa = 11.84GDD368 pKa = 3.71LEE370 pKa = 4.41LGNGGSLLIDD380 pKa = 3.6PTTVTIAQGQGSTDD394 pKa = 3.38NGSDD398 pKa = 2.97ATVFEE403 pKa = 4.3QTLEE407 pKa = 3.99GLLAQGASVYY417 pKa = 10.1IAADD421 pKa = 3.56QAIHH425 pKa = 6.24VADD428 pKa = 5.05LSANGGDD435 pKa = 4.04GILDD439 pKa = 3.71GRR441 pKa = 11.84AGGYY445 pKa = 9.86GGSLTLGIGTQGYY458 pKa = 9.81GGAFGTGTGFSRR470 pKa = 11.84GDD472 pKa = 3.55GVSGGGIFFDD482 pKa = 3.7NTANTIAVDD491 pKa = 3.45GDD493 pKa = 3.77LAILGGYY500 pKa = 6.64TAGEE504 pKa = 4.07LTLGNLIGNSIDD516 pKa = 5.1LEE518 pKa = 4.28AADD521 pKa = 5.59AIHH524 pKa = 6.94ALGSITANNNGQIYY538 pKa = 9.91IDD540 pKa = 3.71STGGDD545 pKa = 3.7VTTHH549 pKa = 6.72ALNGGDD555 pKa = 3.03ITVSAEE561 pKa = 3.96TGALAALGDD570 pKa = 3.82ITISSGTGTVDD581 pKa = 3.74LRR583 pKa = 11.84AGHH586 pKa = 7.22DD587 pKa = 3.65LTVGAITIDD596 pKa = 3.48AGSAPQSYY604 pKa = 10.06GGSIDD609 pKa = 3.48YY610 pKa = 11.16GSIRR614 pKa = 11.84LSADD618 pKa = 3.49DD619 pKa = 4.79GDD621 pKa = 4.54LSVDD625 pKa = 3.3GATLVGDD632 pKa = 4.4LEE634 pKa = 5.22ASLSVAYY641 pKa = 10.27GGTPGLLSITDD652 pKa = 3.8STIEE656 pKa = 4.77GYY658 pKa = 7.79TALYY662 pKa = 9.73AGNGAGITLGGVTINDD678 pKa = 3.36NGLYY682 pKa = 10.6ASVSDD687 pKa = 4.45GGALSADD694 pKa = 3.51NLTASDD700 pKa = 4.43IYY702 pKa = 10.41LTSNSSIDD710 pKa = 3.6TSQGASQAAMQLGNLTATDD729 pKa = 3.35GGLYY733 pKa = 9.51IDD735 pKa = 4.83AGGDD739 pKa = 2.88ITTLHH744 pKa = 6.65NGTALIQGSEE754 pKa = 4.03NVEE757 pKa = 3.93IYY759 pKa = 10.66AGEE762 pKa = 4.44SYY764 pKa = 11.01DD765 pKa = 4.09SEE767 pKa = 4.51TGTTVGGNITLDD779 pKa = 3.74SVHH782 pKa = 6.87AGYY785 pKa = 10.76GIDD788 pKa = 3.99ISNDD792 pKa = 2.95SASYY796 pKa = 9.89YY797 pKa = 10.85GGAFTVGSHH806 pKa = 7.67DD807 pKa = 3.13ITFAHH812 pKa = 6.57LTADD816 pKa = 3.32AGSIYY821 pKa = 9.37VTAEE825 pKa = 3.9RR826 pKa = 11.84NVLATDD832 pKa = 4.12PASSTILATDD842 pKa = 3.6VAYY845 pKa = 10.79LSGEE849 pKa = 3.93NLTIGDD855 pKa = 3.83VSADD859 pKa = 3.65SVDD862 pKa = 3.48LNAYY866 pKa = 9.76GGTINTGAITAFAGNSYY883 pKa = 10.96SYY885 pKa = 11.48GDD887 pKa = 4.54DD888 pKa = 3.16IFFGLHH894 pKa = 6.65DD895 pKa = 4.94LGEE898 pKa = 4.08AHH900 pKa = 6.79GAIGTTGPIRR910 pKa = 11.84VTGGAGSVAIFADD923 pKa = 2.91SGYY926 pKa = 11.05AVSGLNIDD934 pKa = 4.3AGIDD938 pKa = 3.63PVDD941 pKa = 3.63GTRR944 pKa = 11.84GSIEE948 pKa = 4.05FFSSLGSLSVTDD960 pKa = 3.81STLHH964 pKa = 6.42GDD966 pKa = 3.51VTALLLGYY974 pKa = 9.94SDD976 pKa = 3.5QSYY979 pKa = 11.14GGNQLGTLAVGVPTVGDD996 pKa = 3.56DD997 pKa = 3.21KK998 pKa = 11.74LYY1000 pKa = 10.39IDD1002 pKa = 5.11GVAIDD1007 pKa = 4.7GDD1009 pKa = 4.1LVAYY1013 pKa = 9.22TDD1015 pKa = 3.77GGQAIDD1021 pKa = 4.11IGTLTATGEE1030 pKa = 4.2AVIGEE1035 pKa = 4.48YY1036 pKa = 10.77SSVPASSITALDD1048 pKa = 3.27ITAGGIEE1055 pKa = 4.37LGRR1058 pKa = 11.84DD1059 pKa = 3.36FAYY1062 pKa = 10.51GGTVGTDD1069 pKa = 2.85ILVGSLHH1076 pKa = 6.08ATDD1079 pKa = 4.54GDD1081 pKa = 4.11VEE1083 pKa = 4.21ILAGRR1088 pKa = 11.84NIGIISGSTGTVTADD1103 pKa = 2.93QGGVYY1108 pKa = 10.29LSAGTAQIDD1117 pKa = 3.68TGNGYY1122 pKa = 10.08AAGGGDD1128 pKa = 4.05IEE1130 pKa = 5.8IGDD1133 pKa = 3.73LQARR1137 pKa = 11.84QVAIDD1142 pKa = 4.47LYY1144 pKa = 11.01GGQLDD1149 pKa = 3.85TGAITVGGGSDD1160 pKa = 3.74YY1161 pKa = 11.16YY1162 pKa = 10.98DD1163 pKa = 3.4VYY1165 pKa = 11.28FGFHH1169 pKa = 7.01KK1170 pKa = 10.89DD1171 pKa = 3.12SDD1173 pKa = 4.67SIAHH1177 pKa = 6.95IGTIEE1182 pKa = 4.39SISVTDD1188 pKa = 3.8HH1189 pKa = 6.74AGHH1192 pKa = 6.45VYY1194 pKa = 10.14IGSDD1198 pKa = 3.4GDD1200 pKa = 4.15LDD1202 pKa = 3.48IASIGRR1208 pKa = 11.84IDD1210 pKa = 4.17SGSGTDD1216 pKa = 4.03GYY1218 pKa = 11.63GGTTLGSVSIGSEE1231 pKa = 4.0GGDD1234 pKa = 3.17LRR1236 pKa = 11.84VNALDD1241 pKa = 3.93VIGDD1245 pKa = 3.91LSIHH1249 pKa = 4.99MTNVIAHH1256 pKa = 7.3DD1257 pKa = 4.14GNGIALTASGSTSIFANMAAQDD1279 pKa = 3.51VHH1281 pKa = 8.26IDD1283 pKa = 3.19AGRR1286 pKa = 11.84DD1287 pKa = 3.26ISVDD1291 pKa = 4.13DD1292 pKa = 4.43IDD1294 pKa = 5.03AGSDD1298 pKa = 3.48GSVALLAHH1306 pKa = 7.2DD1307 pKa = 4.06GDD1309 pKa = 4.69LYY1311 pKa = 11.44RR1312 pKa = 11.84NGTISTADD1320 pKa = 3.76LSLQANSLSLDD1331 pKa = 4.17PIVLSGSLSLTSNTGGIEE1349 pKa = 3.98LGGAQASSIDD1359 pKa = 3.51LHH1361 pKa = 6.32AANGIDD1367 pKa = 5.14SGDD1370 pKa = 3.85LNAGSSGSIVLVADD1384 pKa = 3.97SGAITTGALTTGNLSVTATSLDD1406 pKa = 3.72FGDD1409 pKa = 3.57VDD1411 pKa = 3.63IAGDD1415 pKa = 4.06LSLTASGGDD1424 pKa = 3.34LSVGNLTAGSVSLSATDD1441 pKa = 3.6ALSVGGQIGSSGATSLNGASIALADD1466 pKa = 4.14QITGSSITIDD1476 pKa = 3.2GGALSGILDD1485 pKa = 4.23LVAGAGGIALDD1496 pKa = 4.06NSSGTVDD1503 pKa = 3.42SASLSSTGAVGIVGPLTATGAIAIDD1528 pKa = 3.95GGSIALADD1536 pKa = 3.76QLSASSITIDD1546 pKa = 3.95GGALSGKK1553 pKa = 9.98LDD1555 pKa = 3.76LQAGPGGIALHH1566 pKa = 6.2NSGSNVTSAKK1576 pKa = 10.06LDD1578 pKa = 3.62SGSKK1582 pKa = 9.93ISIAASLTASGNVTLNAAGSADD1604 pKa = 3.59FANITAHH1611 pKa = 6.89NITIDD1616 pKa = 3.53AGSDD1620 pKa = 3.19STLTHH1625 pKa = 6.48GSVLTAASGKK1635 pKa = 8.98VHH1637 pKa = 7.08LGGPSFSGSSLTVASASLDD1656 pKa = 3.85LASDD1660 pKa = 4.61LYY1662 pKa = 11.7VDD1664 pKa = 4.85ALDD1667 pKa = 4.43IDD1669 pKa = 4.05VSGANGLSLDD1679 pKa = 3.76NVLLSGTSIDD1689 pKa = 4.06LGATHH1694 pKa = 7.86GITLDD1699 pKa = 4.18DD1700 pKa = 3.88LTIDD1704 pKa = 4.0ASQIGLTAGDD1714 pKa = 4.17TLTATDD1720 pKa = 3.78TVLAGNLTATAGGTAQIDD1738 pKa = 3.46NATWQGGTASLEE1750 pKa = 4.09AASLDD1755 pKa = 3.44IGNSQLAYY1763 pKa = 7.84TQSASFITSDD1773 pKa = 3.98GSLNLIDD1780 pKa = 5.07SSFGPISTVQPLAATQSPTVGSGNLSLQSAGDD1812 pKa = 3.67IVLGGSDD1819 pKa = 3.1IRR1821 pKa = 11.84SGRR1824 pKa = 11.84LTLTAAGGILDD1835 pKa = 5.31DD1836 pKa = 5.11GSAANIDD1843 pKa = 3.66VDD1845 pKa = 4.47ALRR1848 pKa = 11.84ANAGTAIDD1856 pKa = 4.19FARR1859 pKa = 11.84SSLIIGTGSSGVAGDD1874 pKa = 3.99SGLLQLLAAQAPQLLPTATAPNGSLHH1900 pKa = 6.09AQSIALGTLAMSGDD1914 pKa = 4.25YY1915 pKa = 11.1LSLTTNALTIGHH1927 pKa = 6.35VSAAPAHH1934 pKa = 6.54LLVQIQPLAATTGIEE1949 pKa = 4.08LSQPTTVAARR1959 pKa = 11.84ALSSPAALNFSDD1971 pKa = 4.05AAAFAPYY1978 pKa = 9.79DD1979 pKa = 3.46ASLAFGGSNYY1989 pKa = 10.7AGDD1992 pKa = 4.0INVDD1996 pKa = 3.46SGVDD2000 pKa = 3.41VANASTNFLFMTSGHH2015 pKa = 5.56VNGHH2019 pKa = 6.39RR2020 pKa = 11.84NLATNGQVIVLSGIVTTEE2038 pKa = 3.7NLSDD2042 pKa = 4.23FEE2044 pKa = 4.82LPLIGEE2050 pKa = 4.24IQSGGLGDD2058 pKa = 4.63TPPLDD2063 pKa = 5.06DD2064 pKa = 5.29NPAAHH2069 pKa = 7.46PDD2071 pKa = 3.81DD2072 pKa = 4.18TSSLITIEE2080 pKa = 4.66DD2081 pKa = 3.8APGDD2085 pKa = 4.03TQCRR2089 pKa = 3.74
Molecular weight: 204.81 kDa
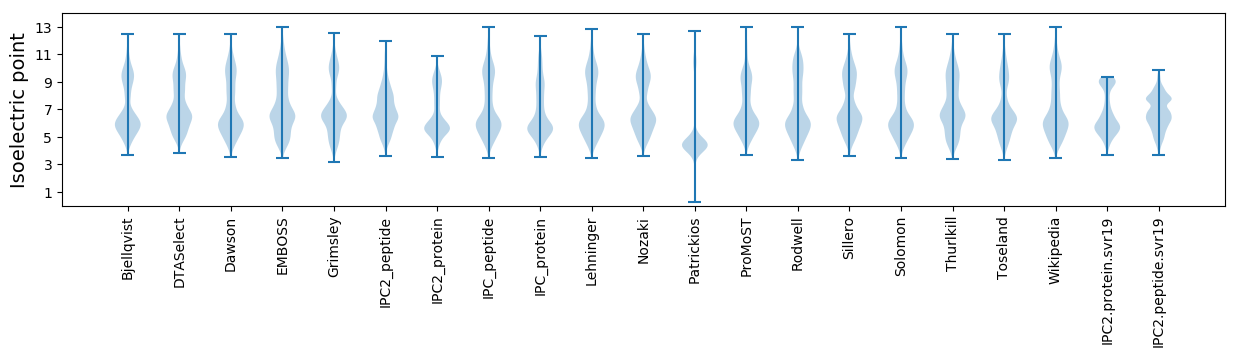
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M2BQS4|A0A6M2BQS4_9GAMM Nuclear transport factor 2 family protein OS=Solimonas terrae OX=1396819 GN=G7Y85_08005 PE=4 SV=1
MM1 pKa = 7.45EE2 pKa = 5.05GSAAIEE8 pKa = 3.9RR9 pKa = 11.84LPSRR13 pKa = 11.84DD14 pKa = 3.05RR15 pKa = 11.84DD16 pKa = 3.42GRR18 pKa = 11.84AVPNGTPRR26 pKa = 11.84QAVAEE31 pKa = 4.31RR32 pKa = 11.84PNLTLVGPHH41 pKa = 5.57TRR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 4.07VRR47 pKa = 11.84PATAKK52 pKa = 6.6TTVYY56 pKa = 10.27QAFGKK61 pKa = 10.42RR62 pKa = 11.84LFDD65 pKa = 4.92IIFSLAFLVAVGSWLFPLLAIAIRR89 pKa = 11.84ANSKK93 pKa = 10.25GPVFFRR99 pKa = 11.84QRR101 pKa = 11.84RR102 pKa = 11.84VGLGGEE108 pKa = 4.3VFTCLKK114 pKa = 9.29FRR116 pKa = 11.84TMAHH120 pKa = 6.38CPNAGFVQAQKK131 pKa = 10.91DD132 pKa = 3.77DD133 pKa = 3.8CRR135 pKa = 11.84ITAVGRR141 pKa = 11.84FLRR144 pKa = 11.84RR145 pKa = 11.84TNLDD149 pKa = 3.34EE150 pKa = 4.32VPQFVNVLVGDD161 pKa = 3.89MSVIGPRR168 pKa = 11.84PHH170 pKa = 7.91VPEE173 pKa = 5.27LDD175 pKa = 3.62GMFKK179 pKa = 10.83DD180 pKa = 5.43LIPAYY185 pKa = 6.88TQRR188 pKa = 11.84NSVKK192 pKa = 10.17PGVSGLAQVSGCRR205 pKa = 11.84GEE207 pKa = 4.19TRR209 pKa = 11.84SVRR212 pKa = 11.84EE213 pKa = 3.52MNHH216 pKa = 5.52RR217 pKa = 11.84VRR219 pKa = 11.84FDD221 pKa = 3.09VFYY224 pKa = 10.84CRR226 pKa = 11.84NVSFAMDD233 pKa = 3.57VKK235 pKa = 10.35LVCLTVLRR243 pKa = 11.84ALRR246 pKa = 11.84GDD248 pKa = 3.31EE249 pKa = 3.73RR250 pKa = 11.84AYY252 pKa = 10.82
MM1 pKa = 7.45EE2 pKa = 5.05GSAAIEE8 pKa = 3.9RR9 pKa = 11.84LPSRR13 pKa = 11.84DD14 pKa = 3.05RR15 pKa = 11.84DD16 pKa = 3.42GRR18 pKa = 11.84AVPNGTPRR26 pKa = 11.84QAVAEE31 pKa = 4.31RR32 pKa = 11.84PNLTLVGPHH41 pKa = 5.57TRR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 4.07VRR47 pKa = 11.84PATAKK52 pKa = 6.6TTVYY56 pKa = 10.27QAFGKK61 pKa = 10.42RR62 pKa = 11.84LFDD65 pKa = 4.92IIFSLAFLVAVGSWLFPLLAIAIRR89 pKa = 11.84ANSKK93 pKa = 10.25GPVFFRR99 pKa = 11.84QRR101 pKa = 11.84RR102 pKa = 11.84VGLGGEE108 pKa = 4.3VFTCLKK114 pKa = 9.29FRR116 pKa = 11.84TMAHH120 pKa = 6.38CPNAGFVQAQKK131 pKa = 10.91DD132 pKa = 3.77DD133 pKa = 3.8CRR135 pKa = 11.84ITAVGRR141 pKa = 11.84FLRR144 pKa = 11.84RR145 pKa = 11.84TNLDD149 pKa = 3.34EE150 pKa = 4.32VPQFVNVLVGDD161 pKa = 3.89MSVIGPRR168 pKa = 11.84PHH170 pKa = 7.91VPEE173 pKa = 5.27LDD175 pKa = 3.62GMFKK179 pKa = 10.83DD180 pKa = 5.43LIPAYY185 pKa = 6.88TQRR188 pKa = 11.84NSVKK192 pKa = 10.17PGVSGLAQVSGCRR205 pKa = 11.84GEE207 pKa = 4.19TRR209 pKa = 11.84SVRR212 pKa = 11.84EE213 pKa = 3.52MNHH216 pKa = 5.52RR217 pKa = 11.84VRR219 pKa = 11.84FDD221 pKa = 3.09VFYY224 pKa = 10.84CRR226 pKa = 11.84NVSFAMDD233 pKa = 3.57VKK235 pKa = 10.35LVCLTVLRR243 pKa = 11.84ALRR246 pKa = 11.84GDD248 pKa = 3.31EE249 pKa = 3.73RR250 pKa = 11.84AYY252 pKa = 10.82
Molecular weight: 28.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1331285 |
37 |
3087 |
338.7 |
36.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.703 ± 0.051 | 0.952 ± 0.013 |
6.001 ± 0.029 | 5.093 ± 0.032 |
3.514 ± 0.021 | 8.385 ± 0.03 |
2.316 ± 0.017 | 4.645 ± 0.025 |
3.017 ± 0.031 | 10.645 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.187 ± 0.018 | 2.587 ± 0.024 |
5.211 ± 0.029 | 3.712 ± 0.024 |
7.408 ± 0.045 | 5.46 ± 0.029 |
5.027 ± 0.032 | 7.143 ± 0.032 |
1.436 ± 0.015 | 2.556 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
