
Apis mellifera associated microvirus 36
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
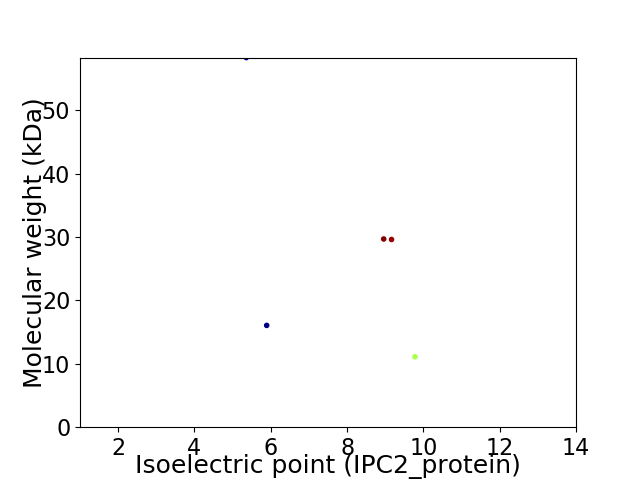
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3S8UUC2|A0A3S8UUC2_9VIRU Uncharacterized protein OS=Apis mellifera associated microvirus 36 OX=2494765 PE=4 SV=1
MM1 pKa = 7.38NLFNQVQIRR10 pKa = 11.84RR11 pKa = 11.84PKK13 pKa = 9.89RR14 pKa = 11.84NKK16 pKa = 9.61FDD18 pKa = 3.91LSHH21 pKa = 6.29EE22 pKa = 4.44KK23 pKa = 10.49KK24 pKa = 10.77LSFNMGDD31 pKa = 5.32LIPIMCQEE39 pKa = 5.36IIPGDD44 pKa = 3.59NFRR47 pKa = 11.84VNTEE51 pKa = 3.59TFMRR55 pKa = 11.84LAPMIAPVMHH65 pKa = 6.62RR66 pKa = 11.84VNVYY70 pKa = 5.97THH72 pKa = 6.06YY73 pKa = 10.7FFVPTRR79 pKa = 11.84IIWDD83 pKa = 3.62EE84 pKa = 3.92FQDD87 pKa = 4.81FITGGRR93 pKa = 11.84EE94 pKa = 4.23GTSMPISPFFGQSTVEE110 pKa = 3.98NARR113 pKa = 11.84GSLLDD118 pKa = 4.0YY119 pKa = 10.19MGLPLWPSGSSNDD132 pKa = 3.68TQISVLPLRR141 pKa = 11.84AYY143 pKa = 7.91QTVWDD148 pKa = 4.8EE149 pKa = 4.1YY150 pKa = 11.32FRR152 pKa = 11.84DD153 pKa = 3.81QNLTPSLDD161 pKa = 3.13ISKK164 pKa = 10.86ASGVMPADD172 pKa = 3.6AEE174 pKa = 4.3LFKK177 pKa = 11.18LLEE180 pKa = 3.95LRR182 pKa = 11.84KK183 pKa = 9.69RR184 pKa = 11.84AWEE187 pKa = 3.7KK188 pKa = 11.28DD189 pKa = 3.61YY190 pKa = 9.27FTSALPWSQRR200 pKa = 11.84GNEE203 pKa = 4.17ILLPMTPEE211 pKa = 3.55VNYY214 pKa = 10.57RR215 pKa = 11.84PTSDD219 pKa = 3.46VYY221 pKa = 11.68YY222 pKa = 10.74EE223 pKa = 5.75DD224 pKa = 3.57GTQANNSSWGIGEE237 pKa = 4.36GPAGEE242 pKa = 4.74DD243 pKa = 3.49GKK245 pKa = 10.79IVPTPGANILQMRR258 pKa = 11.84IEE260 pKa = 4.9NIDD263 pKa = 3.83SIDD266 pKa = 3.92NVTTTINDD274 pKa = 2.95LRR276 pKa = 11.84RR277 pKa = 11.84AVRR280 pKa = 11.84LQEE283 pKa = 3.92WLEE286 pKa = 3.87KK287 pKa = 9.97NARR290 pKa = 11.84GGARR294 pKa = 11.84YY295 pKa = 9.32IEE297 pKa = 4.53QILHH301 pKa = 6.04HH302 pKa = 6.67FGVTSSDD309 pKa = 3.08ARR311 pKa = 11.84LQRR314 pKa = 11.84PEE316 pKa = 3.75YY317 pKa = 10.49LGGGKK322 pKa = 9.57QPVVISEE329 pKa = 4.08VLQTSEE335 pKa = 4.55TLSTPQGTMAGHH347 pKa = 6.45GVSAGTSNGFNRR359 pKa = 11.84SFEE362 pKa = 3.89EE363 pKa = 3.49HH364 pKa = 6.83GYY366 pKa = 10.35VIGVMSVLPRR376 pKa = 11.84TAYY379 pKa = 9.97QDD381 pKa = 3.59GVNKK385 pKa = 9.2MWSRR389 pKa = 11.84TDD391 pKa = 2.89KK392 pKa = 10.86FDD394 pKa = 4.08YY395 pKa = 10.05FWPEE399 pKa = 3.44LANIGEE405 pKa = 4.22QEE407 pKa = 4.22VKK409 pKa = 10.43IKK411 pKa = 10.35EE412 pKa = 4.33LYY414 pKa = 10.67LNPTGAQTNDD424 pKa = 2.76TFGYY428 pKa = 7.52QSRR431 pKa = 11.84YY432 pKa = 10.33AEE434 pKa = 4.25YY435 pKa = 10.17KK436 pKa = 9.33YY437 pKa = 10.4MPSTVHH443 pKa = 6.88GDD445 pKa = 3.03FRR447 pKa = 11.84TNLDD451 pKa = 3.39YY452 pKa = 11.19WHH454 pKa = 7.1MGRR457 pKa = 11.84KK458 pKa = 9.43FSTEE462 pKa = 3.42PVLNEE467 pKa = 4.43DD468 pKa = 5.84FITADD473 pKa = 3.14PTHH476 pKa = 7.26RR477 pKa = 11.84IFAVTDD483 pKa = 3.8PDD485 pKa = 3.47EE486 pKa = 4.78HH487 pKa = 8.32KK488 pKa = 10.75LYY490 pKa = 10.33CQLYY494 pKa = 9.09HH495 pKa = 7.31RR496 pKa = 11.84IDD498 pKa = 3.64ALRR501 pKa = 11.84PMPYY505 pKa = 10.05FGTPTLL511 pKa = 3.89
MM1 pKa = 7.38NLFNQVQIRR10 pKa = 11.84RR11 pKa = 11.84PKK13 pKa = 9.89RR14 pKa = 11.84NKK16 pKa = 9.61FDD18 pKa = 3.91LSHH21 pKa = 6.29EE22 pKa = 4.44KK23 pKa = 10.49KK24 pKa = 10.77LSFNMGDD31 pKa = 5.32LIPIMCQEE39 pKa = 5.36IIPGDD44 pKa = 3.59NFRR47 pKa = 11.84VNTEE51 pKa = 3.59TFMRR55 pKa = 11.84LAPMIAPVMHH65 pKa = 6.62RR66 pKa = 11.84VNVYY70 pKa = 5.97THH72 pKa = 6.06YY73 pKa = 10.7FFVPTRR79 pKa = 11.84IIWDD83 pKa = 3.62EE84 pKa = 3.92FQDD87 pKa = 4.81FITGGRR93 pKa = 11.84EE94 pKa = 4.23GTSMPISPFFGQSTVEE110 pKa = 3.98NARR113 pKa = 11.84GSLLDD118 pKa = 4.0YY119 pKa = 10.19MGLPLWPSGSSNDD132 pKa = 3.68TQISVLPLRR141 pKa = 11.84AYY143 pKa = 7.91QTVWDD148 pKa = 4.8EE149 pKa = 4.1YY150 pKa = 11.32FRR152 pKa = 11.84DD153 pKa = 3.81QNLTPSLDD161 pKa = 3.13ISKK164 pKa = 10.86ASGVMPADD172 pKa = 3.6AEE174 pKa = 4.3LFKK177 pKa = 11.18LLEE180 pKa = 3.95LRR182 pKa = 11.84KK183 pKa = 9.69RR184 pKa = 11.84AWEE187 pKa = 3.7KK188 pKa = 11.28DD189 pKa = 3.61YY190 pKa = 9.27FTSALPWSQRR200 pKa = 11.84GNEE203 pKa = 4.17ILLPMTPEE211 pKa = 3.55VNYY214 pKa = 10.57RR215 pKa = 11.84PTSDD219 pKa = 3.46VYY221 pKa = 11.68YY222 pKa = 10.74EE223 pKa = 5.75DD224 pKa = 3.57GTQANNSSWGIGEE237 pKa = 4.36GPAGEE242 pKa = 4.74DD243 pKa = 3.49GKK245 pKa = 10.79IVPTPGANILQMRR258 pKa = 11.84IEE260 pKa = 4.9NIDD263 pKa = 3.83SIDD266 pKa = 3.92NVTTTINDD274 pKa = 2.95LRR276 pKa = 11.84RR277 pKa = 11.84AVRR280 pKa = 11.84LQEE283 pKa = 3.92WLEE286 pKa = 3.87KK287 pKa = 9.97NARR290 pKa = 11.84GGARR294 pKa = 11.84YY295 pKa = 9.32IEE297 pKa = 4.53QILHH301 pKa = 6.04HH302 pKa = 6.67FGVTSSDD309 pKa = 3.08ARR311 pKa = 11.84LQRR314 pKa = 11.84PEE316 pKa = 3.75YY317 pKa = 10.49LGGGKK322 pKa = 9.57QPVVISEE329 pKa = 4.08VLQTSEE335 pKa = 4.55TLSTPQGTMAGHH347 pKa = 6.45GVSAGTSNGFNRR359 pKa = 11.84SFEE362 pKa = 3.89EE363 pKa = 3.49HH364 pKa = 6.83GYY366 pKa = 10.35VIGVMSVLPRR376 pKa = 11.84TAYY379 pKa = 9.97QDD381 pKa = 3.59GVNKK385 pKa = 9.2MWSRR389 pKa = 11.84TDD391 pKa = 2.89KK392 pKa = 10.86FDD394 pKa = 4.08YY395 pKa = 10.05FWPEE399 pKa = 3.44LANIGEE405 pKa = 4.22QEE407 pKa = 4.22VKK409 pKa = 10.43IKK411 pKa = 10.35EE412 pKa = 4.33LYY414 pKa = 10.67LNPTGAQTNDD424 pKa = 2.76TFGYY428 pKa = 7.52QSRR431 pKa = 11.84YY432 pKa = 10.33AEE434 pKa = 4.25YY435 pKa = 10.17KK436 pKa = 9.33YY437 pKa = 10.4MPSTVHH443 pKa = 6.88GDD445 pKa = 3.03FRR447 pKa = 11.84TNLDD451 pKa = 3.39YY452 pKa = 11.19WHH454 pKa = 7.1MGRR457 pKa = 11.84KK458 pKa = 9.43FSTEE462 pKa = 3.42PVLNEE467 pKa = 4.43DD468 pKa = 5.84FITADD473 pKa = 3.14PTHH476 pKa = 7.26RR477 pKa = 11.84IFAVTDD483 pKa = 3.8PDD485 pKa = 3.47EE486 pKa = 4.78HH487 pKa = 8.32KK488 pKa = 10.75LYY490 pKa = 10.33CQLYY494 pKa = 9.09HH495 pKa = 7.31RR496 pKa = 11.84IDD498 pKa = 3.64ALRR501 pKa = 11.84PMPYY505 pKa = 10.05FGTPTLL511 pKa = 3.89
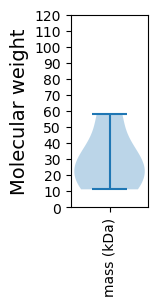
Molecular weight: 58.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q8U647|A0A3Q8U647_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 36 OX=2494765 PE=4 SV=1
MM1 pKa = 6.59YY2 pKa = 10.35QVGVVLKK9 pKa = 7.92FAPYY13 pKa = 10.67SPFNPTSCRR22 pKa = 11.84PTSQRR27 pKa = 11.84HH28 pKa = 4.75AATLIPRR35 pKa = 11.84GVGATIGRR43 pKa = 11.84RR44 pKa = 11.84WAVDD48 pKa = 3.27RR49 pKa = 11.84GRR51 pKa = 11.84KK52 pKa = 8.8IKK54 pKa = 9.62PCKK57 pKa = 9.65GFKK60 pKa = 9.85IGLKK64 pKa = 9.9TNTLPLGAPGRR75 pKa = 11.84TNATCHH81 pKa = 6.68DD82 pKa = 4.13GYY84 pKa = 11.5GRR86 pKa = 11.84DD87 pKa = 4.25GLNPWALEE95 pKa = 3.94APLPLNGII103 pKa = 4.31
MM1 pKa = 6.59YY2 pKa = 10.35QVGVVLKK9 pKa = 7.92FAPYY13 pKa = 10.67SPFNPTSCRR22 pKa = 11.84PTSQRR27 pKa = 11.84HH28 pKa = 4.75AATLIPRR35 pKa = 11.84GVGATIGRR43 pKa = 11.84RR44 pKa = 11.84WAVDD48 pKa = 3.27RR49 pKa = 11.84GRR51 pKa = 11.84KK52 pKa = 8.8IKK54 pKa = 9.62PCKK57 pKa = 9.65GFKK60 pKa = 9.85IGLKK64 pKa = 9.9TNTLPLGAPGRR75 pKa = 11.84TNATCHH81 pKa = 6.68DD82 pKa = 4.13GYY84 pKa = 11.5GRR86 pKa = 11.84DD87 pKa = 4.25GLNPWALEE95 pKa = 3.94APLPLNGII103 pKa = 4.31
Molecular weight: 11.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1270 |
103 |
511 |
254.0 |
28.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.402 ± 1.387 | 0.551 ± 0.261 |
4.803 ± 0.655 | 6.929 ± 1.267 |
3.78 ± 0.708 | 7.087 ± 0.836 |
2.835 ± 0.909 | 5.433 ± 0.534 |
6.772 ± 2.171 | 7.638 ± 0.242 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.913 ± 0.424 | 6.063 ± 1.023 |
5.591 ± 0.939 | 4.252 ± 0.714 |
6.378 ± 0.638 | 5.118 ± 0.497 |
6.457 ± 0.502 | 4.016 ± 0.681 |
1.732 ± 0.341 | 4.252 ± 0.388 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
