
Grapevine virus A (isolate Is 151) (GVA)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Betaflexiviridae; Trivirinae; Vitivirus; Grapevine virus A
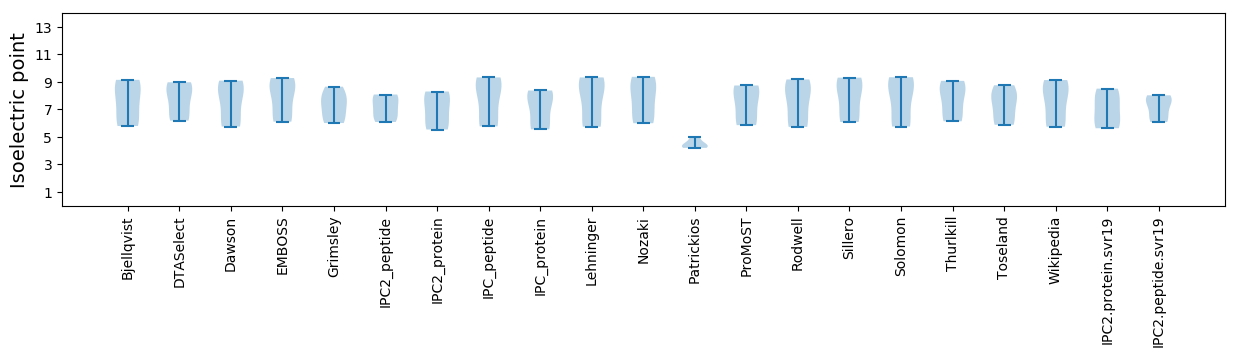
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
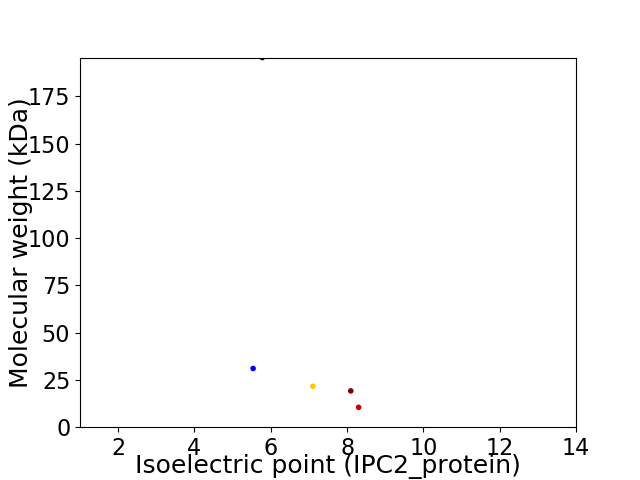
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q67707|CAPSD_GVAIS Capsid protein OS=Grapevine virus A (isolate Is 151) OX=651358 GN=ORF4 PE=4 SV=1
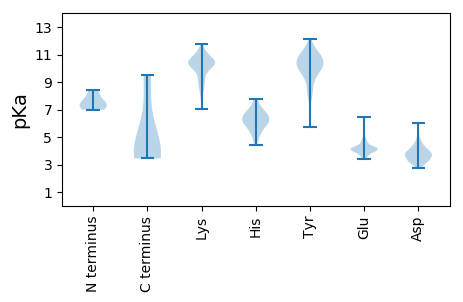
MM1 pKa = 6.99SQEE4 pKa = 3.94GSLGTKK10 pKa = 10.04ASSFEE15 pKa = 3.97PQDD18 pKa = 2.89IKK20 pKa = 11.3VFHH23 pKa = 6.45VKK25 pKa = 10.21RR26 pKa = 11.84STRR29 pKa = 11.84DD30 pKa = 3.19LEE32 pKa = 4.3TLNKK36 pKa = 9.63SLHH39 pKa = 6.32RR40 pKa = 11.84GDD42 pKa = 3.96VYY44 pKa = 10.69NTEE47 pKa = 4.74LIEE50 pKa = 4.25KK51 pKa = 8.5VFPRR55 pKa = 11.84RR56 pKa = 11.84TKK58 pKa = 10.17KK59 pKa = 10.5CVIHH63 pKa = 6.97KK64 pKa = 10.37DD65 pKa = 3.64VIVKK69 pKa = 10.29DD70 pKa = 3.76GRR72 pKa = 11.84VDD74 pKa = 3.69CDD76 pKa = 4.15LDD78 pKa = 4.1IMDD81 pKa = 5.42EE82 pKa = 4.18GLDD85 pKa = 4.39DD86 pKa = 4.0INEE89 pKa = 4.22EE90 pKa = 4.27EE91 pKa = 4.35FPLYY95 pKa = 10.34HH96 pKa = 6.41VGCIVVALMPHH107 pKa = 6.48GKK109 pKa = 9.03NLQGKK114 pKa = 9.05VSVEE118 pKa = 3.99VLDD121 pKa = 3.77TRR123 pKa = 11.84LVDD126 pKa = 3.56GASRR130 pKa = 11.84ISRR133 pKa = 11.84TLMDD137 pKa = 3.24MSKK140 pKa = 10.24PLSACADD147 pKa = 3.56FPGYY151 pKa = 10.31FISTSDD157 pKa = 3.79LLNGYY162 pKa = 6.52TLHH165 pKa = 7.79LSITTTDD172 pKa = 3.39LQFVDD177 pKa = 4.93GVHH180 pKa = 6.45PFSVQLMSIGRR191 pKa = 11.84FCGEE195 pKa = 4.16DD196 pKa = 2.81MKK198 pKa = 10.94TRR200 pKa = 11.84YY201 pKa = 10.26AITEE205 pKa = 4.11TSKK208 pKa = 10.27MLHH211 pKa = 5.24QNILNTEE218 pKa = 4.23GDD220 pKa = 3.75GEE222 pKa = 4.74LIPRR226 pKa = 11.84GVQVQKK232 pKa = 11.16VPDD235 pKa = 3.98TLVMPEE241 pKa = 3.83VFEE244 pKa = 4.5TIKK247 pKa = 11.01KK248 pKa = 10.33FGLKK252 pKa = 9.67TNGTLRR258 pKa = 11.84QEE260 pKa = 4.47GRR262 pKa = 11.84DD263 pKa = 3.22KK264 pKa = 11.44GDD266 pKa = 3.09NRR268 pKa = 11.84RR269 pKa = 11.84VGVGEE274 pKa = 4.19SPTNN278 pKa = 3.44
MM1 pKa = 6.99SQEE4 pKa = 3.94GSLGTKK10 pKa = 10.04ASSFEE15 pKa = 3.97PQDD18 pKa = 2.89IKK20 pKa = 11.3VFHH23 pKa = 6.45VKK25 pKa = 10.21RR26 pKa = 11.84STRR29 pKa = 11.84DD30 pKa = 3.19LEE32 pKa = 4.3TLNKK36 pKa = 9.63SLHH39 pKa = 6.32RR40 pKa = 11.84GDD42 pKa = 3.96VYY44 pKa = 10.69NTEE47 pKa = 4.74LIEE50 pKa = 4.25KK51 pKa = 8.5VFPRR55 pKa = 11.84RR56 pKa = 11.84TKK58 pKa = 10.17KK59 pKa = 10.5CVIHH63 pKa = 6.97KK64 pKa = 10.37DD65 pKa = 3.64VIVKK69 pKa = 10.29DD70 pKa = 3.76GRR72 pKa = 11.84VDD74 pKa = 3.69CDD76 pKa = 4.15LDD78 pKa = 4.1IMDD81 pKa = 5.42EE82 pKa = 4.18GLDD85 pKa = 4.39DD86 pKa = 4.0INEE89 pKa = 4.22EE90 pKa = 4.27EE91 pKa = 4.35FPLYY95 pKa = 10.34HH96 pKa = 6.41VGCIVVALMPHH107 pKa = 6.48GKK109 pKa = 9.03NLQGKK114 pKa = 9.05VSVEE118 pKa = 3.99VLDD121 pKa = 3.77TRR123 pKa = 11.84LVDD126 pKa = 3.56GASRR130 pKa = 11.84ISRR133 pKa = 11.84TLMDD137 pKa = 3.24MSKK140 pKa = 10.24PLSACADD147 pKa = 3.56FPGYY151 pKa = 10.31FISTSDD157 pKa = 3.79LLNGYY162 pKa = 6.52TLHH165 pKa = 7.79LSITTTDD172 pKa = 3.39LQFVDD177 pKa = 4.93GVHH180 pKa = 6.45PFSVQLMSIGRR191 pKa = 11.84FCGEE195 pKa = 4.16DD196 pKa = 2.81MKK198 pKa = 10.94TRR200 pKa = 11.84YY201 pKa = 10.26AITEE205 pKa = 4.11TSKK208 pKa = 10.27MLHH211 pKa = 5.24QNILNTEE218 pKa = 4.23GDD220 pKa = 3.75GEE222 pKa = 4.74LIPRR226 pKa = 11.84GVQVQKK232 pKa = 11.16VPDD235 pKa = 3.98TLVMPEE241 pKa = 3.83VFEE244 pKa = 4.5TIKK247 pKa = 11.01KK248 pKa = 10.33FGLKK252 pKa = 9.67TNGTLRR258 pKa = 11.84QEE260 pKa = 4.47GRR262 pKa = 11.84DD263 pKa = 3.22KK264 pKa = 11.44GDD266 pKa = 3.09NRR268 pKa = 11.84RR269 pKa = 11.84VGVGEE274 pKa = 4.19SPTNN278 pKa = 3.44
Molecular weight: 31.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q67708|VSR_GVAIS RNA silencing suppressor OS=Grapevine virus A (isolate Is 151) OX=651358 GN=ORF5 PE=3 SV=2
MM1 pKa = 8.42DD2 pKa = 5.95DD3 pKa = 3.85PSFLTGRR10 pKa = 11.84STFAKK15 pKa = 9.46RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84RR21 pKa = 11.84MNVCKK26 pKa = 10.46CGAIMHH32 pKa = 6.32NNKK35 pKa = 9.59DD36 pKa = 4.19CKK38 pKa = 10.37SSSISSHH45 pKa = 5.93KK46 pKa = 10.17LDD48 pKa = 3.11RR49 pKa = 11.84LRR51 pKa = 11.84FVKK54 pKa = 10.31EE55 pKa = 3.45GRR57 pKa = 11.84VALTGEE63 pKa = 4.29TPVYY67 pKa = 8.51RR68 pKa = 11.84TWVKK72 pKa = 9.15WVEE75 pKa = 4.28TEE77 pKa = 3.6YY78 pKa = 10.74HH79 pKa = 6.0IYY81 pKa = 10.08IVEE84 pKa = 4.14TSDD87 pKa = 5.92DD88 pKa = 3.61EE89 pKa = 4.72DD90 pKa = 3.59
MM1 pKa = 8.42DD2 pKa = 5.95DD3 pKa = 3.85PSFLTGRR10 pKa = 11.84STFAKK15 pKa = 9.46RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84ARR20 pKa = 11.84RR21 pKa = 11.84MNVCKK26 pKa = 10.46CGAIMHH32 pKa = 6.32NNKK35 pKa = 9.59DD36 pKa = 4.19CKK38 pKa = 10.37SSSISSHH45 pKa = 5.93KK46 pKa = 10.17LDD48 pKa = 3.11RR49 pKa = 11.84LRR51 pKa = 11.84FVKK54 pKa = 10.31EE55 pKa = 3.45GRR57 pKa = 11.84VALTGEE63 pKa = 4.29TPVYY67 pKa = 8.51RR68 pKa = 11.84TWVKK72 pKa = 9.15WVEE75 pKa = 4.28TEE77 pKa = 3.6YY78 pKa = 10.74HH79 pKa = 6.0IYY81 pKa = 10.08IVEE84 pKa = 4.14TSDD87 pKa = 5.92DD88 pKa = 3.61EE89 pKa = 4.72DD90 pKa = 3.59
Molecular weight: 10.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2450 |
90 |
1707 |
490.0 |
55.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.551 ± 1.047 | 1.837 ± 0.232 |
6.286 ± 0.554 | 8.122 ± 1.179 |
3.796 ± 0.083 | 6.939 ± 1.115 |
2.898 ± 0.37 | 5.347 ± 0.141 |
6.939 ± 0.573 | 9.755 ± 0.94 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.776 ± 0.458 | 3.306 ± 0.317 |
3.102 ± 0.36 | 2.694 ± 0.36 |
6.49 ± 0.519 | 7.592 ± 1.12 |
5.347 ± 0.684 | 6.531 ± 0.842 |
0.776 ± 0.319 | 3.918 ± 0.522 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
