
Candidatus Mancarchaeum acidiphilum
Taxonomy: cellular organisms; Archaea; DPANN group; Candidatus Micrarchaeota; Candidatus Mancarchaeum
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
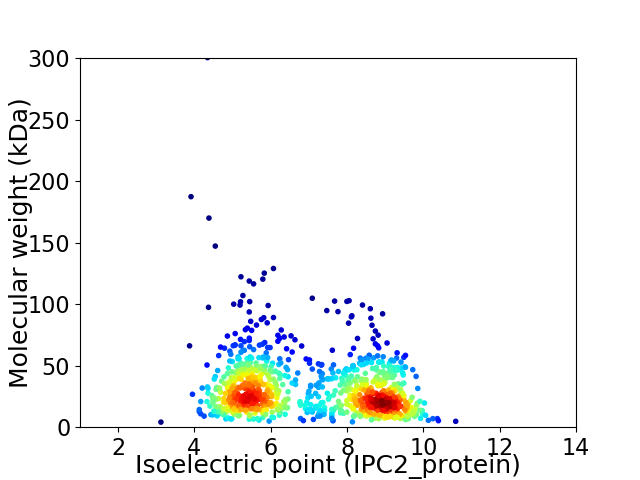
Virtual 2D-PAGE plot for 944 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A218NM28|A0A218NM28_9ARCH AAA+ superfamily ATPase OS=Candidatus Mancarchaeum acidiphilum OX=1920749 GN=Mia14_0183 PE=4 SV=1
MM1 pKa = 7.19TNKK4 pKa = 10.11KK5 pKa = 10.55GFIFTLDD12 pKa = 3.16AVFALIVAAAAISILLYY29 pKa = 9.95IHH31 pKa = 6.91FIAPISYY38 pKa = 9.65QSSISQVEE46 pKa = 4.28DD47 pKa = 3.55DD48 pKa = 4.63LQVLTSTPLGGYY60 pKa = 10.27LSALDD65 pKa = 4.41NPFSGYY71 pKa = 7.57TRR73 pKa = 11.84SYY75 pKa = 8.74NTFGAASFDD84 pKa = 3.99GYY86 pKa = 11.45NSYY89 pKa = 10.57ISSQLSVPAAEE100 pKa = 4.14NLTYY104 pKa = 10.66VAWIYY109 pKa = 11.19LNYY112 pKa = 10.17IPDD115 pKa = 3.74NNAGIISAVSSYY127 pKa = 11.38NYY129 pKa = 10.31GSMSIFLNGNCNIQVLVTNNTGNTFADD156 pKa = 4.04DD157 pKa = 3.79SPYY160 pKa = 10.48CLKK163 pKa = 10.19PHH165 pKa = 6.85RR166 pKa = 11.84WYY168 pKa = 10.82QVVATEE174 pKa = 4.66NEE176 pKa = 4.75SISQDD181 pKa = 2.88GKK183 pKa = 11.25VIGGNYY189 pKa = 7.91TLYY192 pKa = 10.57INGEE196 pKa = 3.92WNLTQKK202 pKa = 10.62FSGALEE208 pKa = 4.27GATGFNIGLFSYY220 pKa = 10.16QGAYY224 pKa = 9.48FPGYY228 pKa = 9.12ISNVQIYY235 pKa = 9.66GSRR238 pKa = 11.84LTRR241 pKa = 11.84TQVSSLYY248 pKa = 10.72DD249 pKa = 3.82EE250 pKa = 4.61GTMGIPLRR258 pKa = 11.84SSNLLEE264 pKa = 4.18WYY266 pKa = 8.98PLNSEE271 pKa = 4.33SNPNVFKK278 pKa = 10.74SYY280 pKa = 10.87NVTYY284 pKa = 10.47FNTGDD289 pKa = 3.24ISIGNVNLTEE299 pKa = 4.12NQSMLSAIADD309 pKa = 4.29LYY311 pKa = 11.66LNDD314 pKa = 4.07QAPEE318 pKa = 3.6ADD320 pKa = 3.36IIMDD324 pKa = 3.88NLYY327 pKa = 10.59HH328 pKa = 7.18NEE330 pKa = 3.83DD331 pKa = 3.02TGIYY335 pKa = 9.75INSTYY340 pKa = 11.17APDD343 pKa = 3.79LKK345 pKa = 10.84VAYY348 pKa = 9.97FDD350 pKa = 3.78GTAGFYY356 pKa = 10.83APNFTYY362 pKa = 10.11PGKK365 pKa = 10.43NYY367 pKa = 9.71TINQWFYY374 pKa = 11.36LSQADD379 pKa = 4.51DD380 pKa = 3.67PTSGVGGGQSPIVDD394 pKa = 4.21IYY396 pKa = 11.24NGSANGGIGGSQNLDD411 pKa = 3.71FGGTWAGGSSNNFGIGEE428 pKa = 4.29DD429 pKa = 3.82WPSDD433 pKa = 3.26WEE435 pKa = 4.44FCSTNAGTVFPNKK448 pKa = 8.67WYY450 pKa = 9.31NAVVSVVNYY459 pKa = 9.52TNVTIYY465 pKa = 11.12LNGVEE470 pKa = 5.35AKK472 pKa = 10.3NCLLTVPISGMEE484 pKa = 3.99YY485 pKa = 7.32PTLGIGDD492 pKa = 4.18NPPGGNEE499 pKa = 3.9LATAAISNVQVYY511 pKa = 10.55DD512 pKa = 3.52SAVNANQAVTLYY524 pKa = 10.82DD525 pKa = 3.35EE526 pKa = 5.92GIAGEE531 pKa = 4.38PVSSSVIGWYY541 pKa = 9.49PLLGNFNDD549 pKa = 4.11YY550 pKa = 11.21SGIGHH555 pKa = 6.5SGFGGNGGSLNPDD568 pKa = 2.53NVRR571 pKa = 11.84FIEE574 pKa = 4.89SNFTPPGFASSDD586 pKa = 3.67IISQASSPEE595 pKa = 3.86LLNVNGTMNVYY606 pKa = 9.9NVSVLSWGG614 pKa = 3.47
MM1 pKa = 7.19TNKK4 pKa = 10.11KK5 pKa = 10.55GFIFTLDD12 pKa = 3.16AVFALIVAAAAISILLYY29 pKa = 9.95IHH31 pKa = 6.91FIAPISYY38 pKa = 9.65QSSISQVEE46 pKa = 4.28DD47 pKa = 3.55DD48 pKa = 4.63LQVLTSTPLGGYY60 pKa = 10.27LSALDD65 pKa = 4.41NPFSGYY71 pKa = 7.57TRR73 pKa = 11.84SYY75 pKa = 8.74NTFGAASFDD84 pKa = 3.99GYY86 pKa = 11.45NSYY89 pKa = 10.57ISSQLSVPAAEE100 pKa = 4.14NLTYY104 pKa = 10.66VAWIYY109 pKa = 11.19LNYY112 pKa = 10.17IPDD115 pKa = 3.74NNAGIISAVSSYY127 pKa = 11.38NYY129 pKa = 10.31GSMSIFLNGNCNIQVLVTNNTGNTFADD156 pKa = 4.04DD157 pKa = 3.79SPYY160 pKa = 10.48CLKK163 pKa = 10.19PHH165 pKa = 6.85RR166 pKa = 11.84WYY168 pKa = 10.82QVVATEE174 pKa = 4.66NEE176 pKa = 4.75SISQDD181 pKa = 2.88GKK183 pKa = 11.25VIGGNYY189 pKa = 7.91TLYY192 pKa = 10.57INGEE196 pKa = 3.92WNLTQKK202 pKa = 10.62FSGALEE208 pKa = 4.27GATGFNIGLFSYY220 pKa = 10.16QGAYY224 pKa = 9.48FPGYY228 pKa = 9.12ISNVQIYY235 pKa = 9.66GSRR238 pKa = 11.84LTRR241 pKa = 11.84TQVSSLYY248 pKa = 10.72DD249 pKa = 3.82EE250 pKa = 4.61GTMGIPLRR258 pKa = 11.84SSNLLEE264 pKa = 4.18WYY266 pKa = 8.98PLNSEE271 pKa = 4.33SNPNVFKK278 pKa = 10.74SYY280 pKa = 10.87NVTYY284 pKa = 10.47FNTGDD289 pKa = 3.24ISIGNVNLTEE299 pKa = 4.12NQSMLSAIADD309 pKa = 4.29LYY311 pKa = 11.66LNDD314 pKa = 4.07QAPEE318 pKa = 3.6ADD320 pKa = 3.36IIMDD324 pKa = 3.88NLYY327 pKa = 10.59HH328 pKa = 7.18NEE330 pKa = 3.83DD331 pKa = 3.02TGIYY335 pKa = 9.75INSTYY340 pKa = 11.17APDD343 pKa = 3.79LKK345 pKa = 10.84VAYY348 pKa = 9.97FDD350 pKa = 3.78GTAGFYY356 pKa = 10.83APNFTYY362 pKa = 10.11PGKK365 pKa = 10.43NYY367 pKa = 9.71TINQWFYY374 pKa = 11.36LSQADD379 pKa = 4.51DD380 pKa = 3.67PTSGVGGGQSPIVDD394 pKa = 4.21IYY396 pKa = 11.24NGSANGGIGGSQNLDD411 pKa = 3.71FGGTWAGGSSNNFGIGEE428 pKa = 4.29DD429 pKa = 3.82WPSDD433 pKa = 3.26WEE435 pKa = 4.44FCSTNAGTVFPNKK448 pKa = 8.67WYY450 pKa = 9.31NAVVSVVNYY459 pKa = 9.52TNVTIYY465 pKa = 11.12LNGVEE470 pKa = 5.35AKK472 pKa = 10.3NCLLTVPISGMEE484 pKa = 3.99YY485 pKa = 7.32PTLGIGDD492 pKa = 4.18NPPGGNEE499 pKa = 3.9LATAAISNVQVYY511 pKa = 10.55DD512 pKa = 3.52SAVNANQAVTLYY524 pKa = 10.82DD525 pKa = 3.35EE526 pKa = 5.92GIAGEE531 pKa = 4.38PVSSSVIGWYY541 pKa = 9.49PLLGNFNDD549 pKa = 4.11YY550 pKa = 11.21SGIGHH555 pKa = 6.5SGFGGNGGSLNPDD568 pKa = 2.53NVRR571 pKa = 11.84FIEE574 pKa = 4.89SNFTPPGFASSDD586 pKa = 3.67IISQASSPEE595 pKa = 3.86LLNVNGTMNVYY606 pKa = 9.9NVSVLSWGG614 pKa = 3.47
Molecular weight: 66.16 kDa
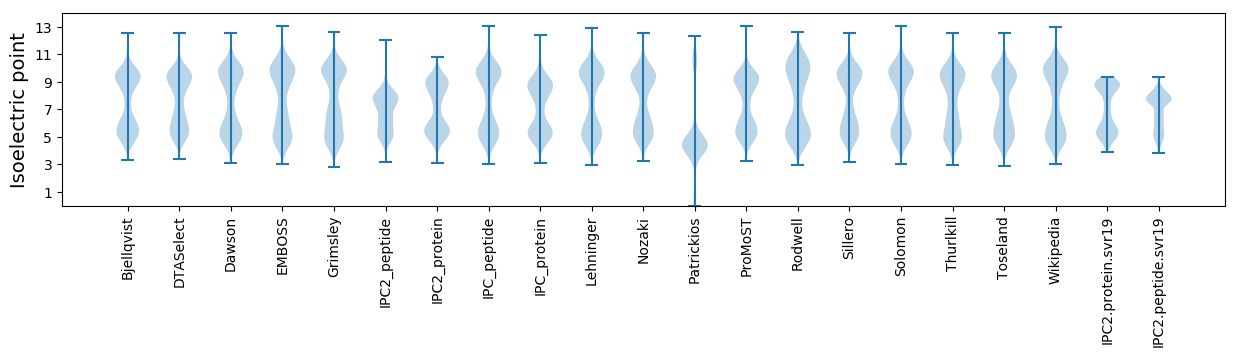
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A218NNS2|A0A218NNS2_9ARCH Chromosome segregation ATPase OS=Candidatus Mancarchaeum acidiphilum OX=1920749 GN=Mia14_0839 PE=4 SV=1
MM1 pKa = 7.57GSIPRR6 pKa = 11.84KK7 pKa = 7.15WKK9 pKa = 10.06KK10 pKa = 9.08KK11 pKa = 10.29GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 8.72WKK19 pKa = 8.65KK20 pKa = 10.1KK21 pKa = 7.96RR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 9.05RR25 pKa = 11.84LKK27 pKa = 8.75RR28 pKa = 11.84AQKK31 pKa = 10.23RR32 pKa = 11.84RR33 pKa = 11.84VGEE36 pKa = 3.92LL37 pKa = 3.03
MM1 pKa = 7.57GSIPRR6 pKa = 11.84KK7 pKa = 7.15WKK9 pKa = 10.06KK10 pKa = 9.08KK11 pKa = 10.29GRR13 pKa = 11.84MRR15 pKa = 11.84WKK17 pKa = 8.72WKK19 pKa = 8.65KK20 pKa = 10.1KK21 pKa = 7.96RR22 pKa = 11.84RR23 pKa = 11.84KK24 pKa = 9.05RR25 pKa = 11.84LKK27 pKa = 8.75RR28 pKa = 11.84AQKK31 pKa = 10.23RR32 pKa = 11.84RR33 pKa = 11.84VGEE36 pKa = 3.92LL37 pKa = 3.03
Molecular weight: 4.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
277313 |
35 |
2805 |
293.8 |
32.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.234 ± 0.076 | 0.675 ± 0.028 |
5.387 ± 0.076 | 6.324 ± 0.095 |
4.351 ± 0.067 | 6.562 ± 0.082 |
1.378 ± 0.028 | 8.965 ± 0.091 |
8.491 ± 0.111 | 9.002 ± 0.1 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.608 ± 0.037 | 5.89 ± 0.108 |
3.453 ± 0.051 | 2.154 ± 0.034 |
3.708 ± 0.075 | 8.457 ± 0.11 |
4.784 ± 0.105 | 6.095 ± 0.064 |
0.74 ± 0.025 | 4.739 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
