
Diaporthe ampelina
Taxonomy:
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
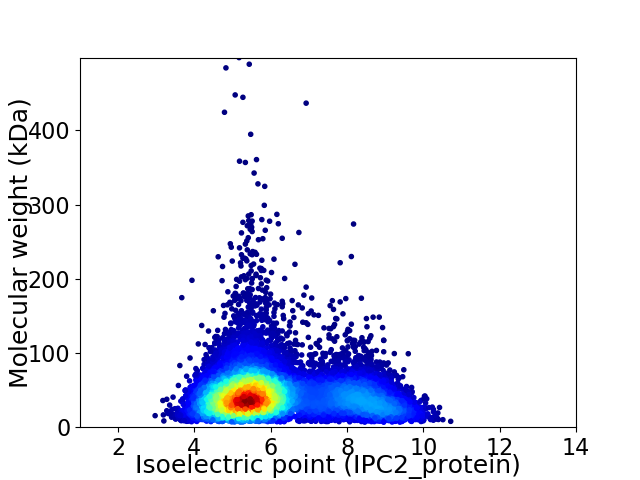
Virtual 2D-PAGE plot for 10703 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0G2HBB3|A0A0G2HBB3_9PEZI Putative mfs maltose OS=Diaporthe ampelina OX=1214573 GN=UCDDA912_g07560 PE=3 SV=1
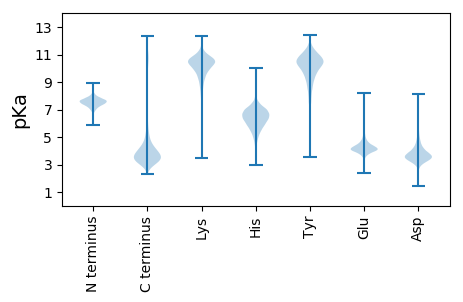
MM1 pKa = 7.45RR2 pKa = 11.84RR3 pKa = 11.84SIFSKK8 pKa = 10.55RR9 pKa = 11.84QDD11 pKa = 2.97ATTLEE16 pKa = 4.27NQQSGTSYY24 pKa = 10.56TIDD27 pKa = 3.44IEE29 pKa = 4.53IGTPPQPITLIIDD42 pKa = 3.81TGSSEE47 pKa = 4.55LWVNPTCEE55 pKa = 3.84TSGQPEE61 pKa = 4.27YY62 pKa = 11.0CEE64 pKa = 4.53SFSQFDD70 pKa = 3.73YY71 pKa = 10.12TASSTINDD79 pKa = 3.53TGYY82 pKa = 11.62SNMLAYY88 pKa = 10.18GKK90 pKa = 10.96GNVTIEE96 pKa = 4.12YY97 pKa = 8.2VTDD100 pKa = 3.62VVSIGSASITDD111 pKa = 3.51QIFGVGFEE119 pKa = 5.0SYY121 pKa = 10.53DD122 pKa = 3.34IPLGILGLAPPTDD135 pKa = 3.97SDD137 pKa = 3.83PLYY140 pKa = 10.71TYY142 pKa = 11.2VLDD145 pKa = 3.93NMVDD149 pKa = 3.05QGLIDD154 pKa = 3.71SRR156 pKa = 11.84AFSLDD161 pKa = 3.1LRR163 pKa = 11.84DD164 pKa = 3.39VDD166 pKa = 4.69SPDD169 pKa = 3.08GSVIFGGVDD178 pKa = 2.76TGKK181 pKa = 10.53FVGEE185 pKa = 3.98LQKK188 pKa = 11.2CPILDD193 pKa = 4.26PLDD196 pKa = 4.13TPSGADD202 pKa = 3.5RR203 pKa = 11.84YY204 pKa = 9.45WIYY207 pKa = 10.33LTGLGMTLPSGEE219 pKa = 4.49SGLIAEE225 pKa = 4.93GEE227 pKa = 4.28LPVFLDD233 pKa = 3.52SGGTMTRR240 pKa = 11.84LPAEE244 pKa = 4.04VFEE247 pKa = 4.79AVGSVFPGAQYY258 pKa = 11.08DD259 pKa = 4.0SEE261 pKa = 4.46SGYY264 pKa = 11.0FLVDD268 pKa = 3.43CDD270 pKa = 4.33VGDD273 pKa = 3.83SSGSVDD279 pKa = 4.43FVFGDD284 pKa = 3.64KK285 pKa = 10.61VISVAFDD292 pKa = 3.37DD293 pKa = 5.39FIWRR297 pKa = 11.84VPSVEE302 pKa = 4.19DD303 pKa = 3.51TCVLGILADD312 pKa = 4.23DD313 pKa = 4.77GEE315 pKa = 4.71FTRR318 pKa = 11.84ATTGGKK324 pKa = 8.45QRR326 pKa = 11.84EE327 pKa = 4.21ADD329 pKa = 3.44TTFAEE334 pKa = 4.65EE335 pKa = 4.33PVLGDD340 pKa = 3.31TFLRR344 pKa = 11.84AAYY347 pKa = 9.66VVYY350 pKa = 10.7DD351 pKa = 3.36QDD353 pKa = 4.09NRR355 pKa = 11.84NLHH358 pKa = 6.37LAQAANCGASLVAISTGLDD377 pKa = 3.11AVPSVTGDD385 pKa = 3.54CTATAPEE392 pKa = 4.0AVLTGSLTATEE403 pKa = 4.62APSTITSGPGGLTSAIAPGPGGTQTGDD430 pKa = 2.97RR431 pKa = 11.84VASSLCLTCKK441 pKa = 10.52NSATGAAHH449 pKa = 7.44PSTTTRR455 pKa = 11.84TANEE459 pKa = 3.98GAMEE463 pKa = 4.41TGSPLAAVGAMAMAALLII481 pKa = 4.38
MM1 pKa = 7.45RR2 pKa = 11.84RR3 pKa = 11.84SIFSKK8 pKa = 10.55RR9 pKa = 11.84QDD11 pKa = 2.97ATTLEE16 pKa = 4.27NQQSGTSYY24 pKa = 10.56TIDD27 pKa = 3.44IEE29 pKa = 4.53IGTPPQPITLIIDD42 pKa = 3.81TGSSEE47 pKa = 4.55LWVNPTCEE55 pKa = 3.84TSGQPEE61 pKa = 4.27YY62 pKa = 11.0CEE64 pKa = 4.53SFSQFDD70 pKa = 3.73YY71 pKa = 10.12TASSTINDD79 pKa = 3.53TGYY82 pKa = 11.62SNMLAYY88 pKa = 10.18GKK90 pKa = 10.96GNVTIEE96 pKa = 4.12YY97 pKa = 8.2VTDD100 pKa = 3.62VVSIGSASITDD111 pKa = 3.51QIFGVGFEE119 pKa = 5.0SYY121 pKa = 10.53DD122 pKa = 3.34IPLGILGLAPPTDD135 pKa = 3.97SDD137 pKa = 3.83PLYY140 pKa = 10.71TYY142 pKa = 11.2VLDD145 pKa = 3.93NMVDD149 pKa = 3.05QGLIDD154 pKa = 3.71SRR156 pKa = 11.84AFSLDD161 pKa = 3.1LRR163 pKa = 11.84DD164 pKa = 3.39VDD166 pKa = 4.69SPDD169 pKa = 3.08GSVIFGGVDD178 pKa = 2.76TGKK181 pKa = 10.53FVGEE185 pKa = 3.98LQKK188 pKa = 11.2CPILDD193 pKa = 4.26PLDD196 pKa = 4.13TPSGADD202 pKa = 3.5RR203 pKa = 11.84YY204 pKa = 9.45WIYY207 pKa = 10.33LTGLGMTLPSGEE219 pKa = 4.49SGLIAEE225 pKa = 4.93GEE227 pKa = 4.28LPVFLDD233 pKa = 3.52SGGTMTRR240 pKa = 11.84LPAEE244 pKa = 4.04VFEE247 pKa = 4.79AVGSVFPGAQYY258 pKa = 11.08DD259 pKa = 4.0SEE261 pKa = 4.46SGYY264 pKa = 11.0FLVDD268 pKa = 3.43CDD270 pKa = 4.33VGDD273 pKa = 3.83SSGSVDD279 pKa = 4.43FVFGDD284 pKa = 3.64KK285 pKa = 10.61VISVAFDD292 pKa = 3.37DD293 pKa = 5.39FIWRR297 pKa = 11.84VPSVEE302 pKa = 4.19DD303 pKa = 3.51TCVLGILADD312 pKa = 4.23DD313 pKa = 4.77GEE315 pKa = 4.71FTRR318 pKa = 11.84ATTGGKK324 pKa = 8.45QRR326 pKa = 11.84EE327 pKa = 4.21ADD329 pKa = 3.44TTFAEE334 pKa = 4.65EE335 pKa = 4.33PVLGDD340 pKa = 3.31TFLRR344 pKa = 11.84AAYY347 pKa = 9.66VVYY350 pKa = 10.7DD351 pKa = 3.36QDD353 pKa = 4.09NRR355 pKa = 11.84NLHH358 pKa = 6.37LAQAANCGASLVAISTGLDD377 pKa = 3.11AVPSVTGDD385 pKa = 3.54CTATAPEE392 pKa = 4.0AVLTGSLTATEE403 pKa = 4.62APSTITSGPGGLTSAIAPGPGGTQTGDD430 pKa = 2.97RR431 pKa = 11.84VASSLCLTCKK441 pKa = 10.52NSATGAAHH449 pKa = 7.44PSTTTRR455 pKa = 11.84TANEE459 pKa = 3.98GAMEE463 pKa = 4.41TGSPLAAVGAMAMAALLII481 pKa = 4.38
Molecular weight: 50.15 kDa
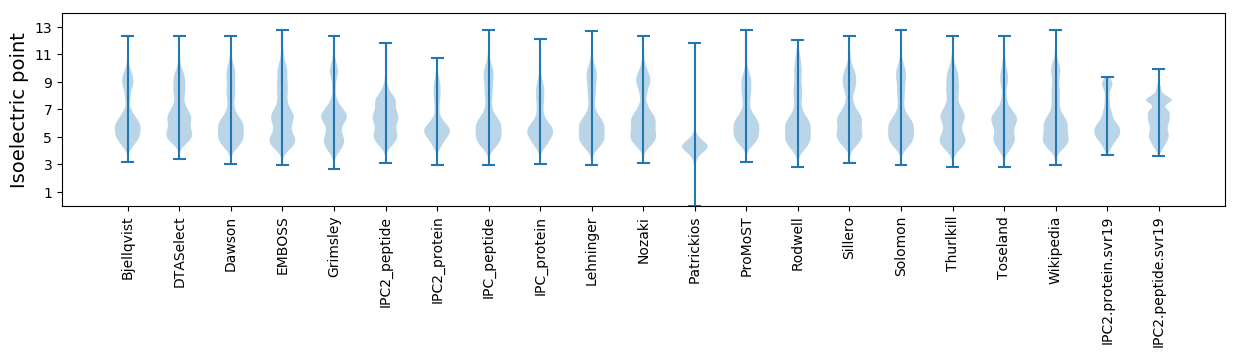
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0G2F7L8|A0A0G2F7L8_9PEZI Probable dipeptidyl-aminopeptidase B OS=Diaporthe ampelina OX=1214573 GN=UCDDA912_g09347 PE=3 SV=1
MM1 pKa = 7.58EE2 pKa = 4.86AQKK5 pKa = 8.88PTQQASLAFRR15 pKa = 11.84PRR17 pKa = 11.84IKK19 pKa = 9.9TSLRR23 pKa = 11.84QRR25 pKa = 11.84FTKK28 pKa = 10.57QKK30 pKa = 10.75DD31 pKa = 3.63PAPKK35 pKa = 10.22APPQDD40 pKa = 3.26QQVAQQSRR48 pKa = 11.84QQNHH52 pKa = 5.29EE53 pKa = 3.69AVATHH58 pKa = 6.19FHH60 pKa = 6.87HH61 pKa = 7.31FPDD64 pKa = 4.54LPTEE68 pKa = 3.97LRR70 pKa = 11.84LQIWAEE76 pKa = 3.48AARR79 pKa = 11.84YY80 pKa = 8.39KK81 pKa = 10.42RR82 pKa = 11.84YY83 pKa = 10.08VVLEE87 pKa = 4.26PPCNSAAACARR98 pKa = 11.84LFLMAKK104 pKa = 9.84RR105 pKa = 11.84YY106 pKa = 9.23GGPGCARR113 pKa = 11.84GRR115 pKa = 11.84YY116 pKa = 8.65RR117 pKa = 11.84PPAWTSRR124 pKa = 11.84TPPPALLAVSTEE136 pKa = 3.67ARR138 pKa = 11.84AVALGTWQRR147 pKa = 11.84AFGYY151 pKa = 10.02GVFPATVMRR160 pKa = 11.84GMHH163 pKa = 6.86RR164 pKa = 11.84LTDD167 pKa = 3.29RR168 pKa = 11.84SEE170 pKa = 4.01FMNGLKK176 pKa = 10.47SLTIEE181 pKa = 4.46IRR183 pKa = 11.84DD184 pKa = 3.65NN185 pKa = 3.48
MM1 pKa = 7.58EE2 pKa = 4.86AQKK5 pKa = 8.88PTQQASLAFRR15 pKa = 11.84PRR17 pKa = 11.84IKK19 pKa = 9.9TSLRR23 pKa = 11.84QRR25 pKa = 11.84FTKK28 pKa = 10.57QKK30 pKa = 10.75DD31 pKa = 3.63PAPKK35 pKa = 10.22APPQDD40 pKa = 3.26QQVAQQSRR48 pKa = 11.84QQNHH52 pKa = 5.29EE53 pKa = 3.69AVATHH58 pKa = 6.19FHH60 pKa = 6.87HH61 pKa = 7.31FPDD64 pKa = 4.54LPTEE68 pKa = 3.97LRR70 pKa = 11.84LQIWAEE76 pKa = 3.48AARR79 pKa = 11.84YY80 pKa = 8.39KK81 pKa = 10.42RR82 pKa = 11.84YY83 pKa = 10.08VVLEE87 pKa = 4.26PPCNSAAACARR98 pKa = 11.84LFLMAKK104 pKa = 9.84RR105 pKa = 11.84YY106 pKa = 9.23GGPGCARR113 pKa = 11.84GRR115 pKa = 11.84YY116 pKa = 8.65RR117 pKa = 11.84PPAWTSRR124 pKa = 11.84TPPPALLAVSTEE136 pKa = 3.67ARR138 pKa = 11.84AVALGTWQRR147 pKa = 11.84AFGYY151 pKa = 10.02GVFPATVMRR160 pKa = 11.84GMHH163 pKa = 6.86RR164 pKa = 11.84LTDD167 pKa = 3.29RR168 pKa = 11.84SEE170 pKa = 4.01FMNGLKK176 pKa = 10.47SLTIEE181 pKa = 4.46IRR183 pKa = 11.84DD184 pKa = 3.65NN185 pKa = 3.48
Molecular weight: 20.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4723684 |
66 |
4584 |
441.3 |
48.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.528 ± 0.025 | 1.123 ± 0.008 |
6.105 ± 0.017 | 6.221 ± 0.021 |
3.764 ± 0.014 | 7.641 ± 0.024 |
2.234 ± 0.01 | 4.594 ± 0.015 |
4.84 ± 0.023 | 8.799 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.195 ± 0.009 | 3.574 ± 0.012 |
5.576 ± 0.02 | 3.843 ± 0.015 |
5.892 ± 0.022 | 7.418 ± 0.021 |
5.76 ± 0.015 | 6.574 ± 0.019 |
1.523 ± 0.01 | 2.791 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
