
Sweet potato collusive virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Caulimoviridae; Cavemovirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
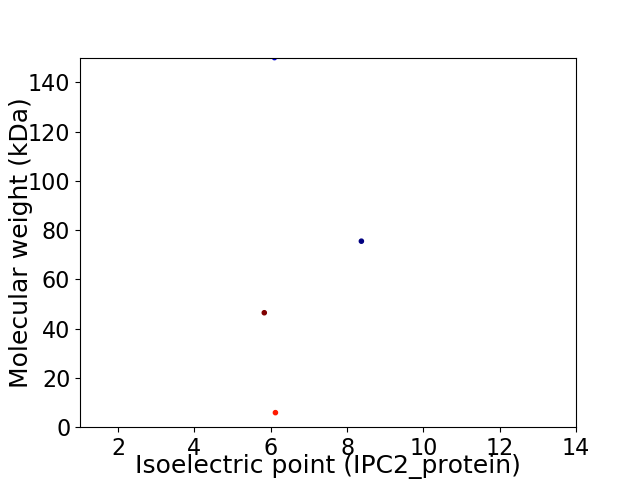
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|F2XXY6|F2XXY6_9VIRU Uncharacterized protein OS=Sweet potato collusive virus OX=930168 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 6.09KK3 pKa = 9.58LTSLLISCPEE13 pKa = 4.13MEE15 pKa = 5.63QMMQQLLAEE24 pKa = 4.24MQKK27 pKa = 11.14LNATVEE33 pKa = 4.4KK34 pKa = 9.97QNSRR38 pKa = 11.84IEE40 pKa = 3.94QLEE43 pKa = 4.06NEE45 pKa = 4.33LEE47 pKa = 4.11KK48 pKa = 11.24EE49 pKa = 4.07KK50 pKa = 10.96GKK52 pKa = 11.02KK53 pKa = 9.43PLTEE57 pKa = 3.76EE58 pKa = 4.39KK59 pKa = 10.25IISEE63 pKa = 4.21KK64 pKa = 10.56EE65 pKa = 3.76LEE67 pKa = 4.15KK68 pKa = 10.91EE69 pKa = 5.34KK70 pKa = 10.74EE71 pKa = 4.07ISWKK75 pKa = 10.88AKK77 pKa = 7.42IKK79 pKa = 10.54KK80 pKa = 9.85YY81 pKa = 11.03GEE83 pKa = 4.14ASSKK87 pKa = 9.13EE88 pKa = 3.82EE89 pKa = 4.16KK90 pKa = 10.14IIEE93 pKa = 4.06KK94 pKa = 10.55DD95 pKa = 3.04VWQKK99 pKa = 11.21PKK101 pKa = 9.27GTHH104 pKa = 5.61IGSIYY109 pKa = 8.26VTEE112 pKa = 5.2FVRR115 pKa = 11.84LMNYY119 pKa = 10.09LNSRR123 pKa = 11.84HH124 pKa = 6.25ANLNEE129 pKa = 3.96IYY131 pKa = 10.83SLTDD135 pKa = 3.29YY136 pKa = 11.37NKK138 pKa = 10.6LVADD142 pKa = 4.01KK143 pKa = 10.03NTDD146 pKa = 2.97EE147 pKa = 4.2RR148 pKa = 11.84LIRR151 pKa = 11.84AAYY154 pKa = 8.85QHH156 pKa = 6.46GLLHH160 pKa = 4.79VHH162 pKa = 6.58YY163 pKa = 9.9IEE165 pKa = 4.93SANQIKK171 pKa = 9.78MYY173 pKa = 10.87DD174 pKa = 3.45EE175 pKa = 5.58GIVNAYY181 pKa = 10.19LKK183 pKa = 10.57LAQLTKK189 pKa = 10.66AKK191 pKa = 10.0CIYY194 pKa = 9.99IRR196 pKa = 11.84FYY198 pKa = 10.47TAFAEE203 pKa = 4.55VTKK206 pKa = 10.67EE207 pKa = 3.83GIIPKK212 pKa = 9.86IEE214 pKa = 4.5AIKK217 pKa = 10.82LGITYY222 pKa = 9.79EE223 pKa = 4.17KK224 pKa = 10.09IQYY227 pKa = 8.71DD228 pKa = 3.66VCEE231 pKa = 4.03QEE233 pKa = 4.93LFEE236 pKa = 4.82EE237 pKa = 4.34NRR239 pKa = 11.84VHH241 pKa = 7.89DD242 pKa = 4.36FLRR245 pKa = 11.84HH246 pKa = 5.06KK247 pKa = 10.43KK248 pKa = 10.53AIGLLTIRR256 pKa = 11.84RR257 pKa = 11.84EE258 pKa = 4.22LEE260 pKa = 3.95STEE263 pKa = 3.93GNIWVYY269 pKa = 8.98EE270 pKa = 3.9NRR272 pKa = 11.84NNRR275 pKa = 11.84IIYY278 pKa = 9.81APSRR282 pKa = 11.84ARR284 pKa = 11.84KK285 pKa = 9.0EE286 pKa = 3.97EE287 pKa = 3.95VKK289 pKa = 10.66KK290 pKa = 11.08VVAAWQQKK298 pKa = 9.39IYY300 pKa = 9.98TPEE303 pKa = 4.02RR304 pKa = 11.84NIPDD308 pKa = 3.82CGITAPCISEE318 pKa = 4.07PCLKK322 pKa = 10.13ILCEE326 pKa = 3.91LSKK329 pKa = 11.02NQLVVKK335 pKa = 8.42HH336 pKa = 4.65QCKK339 pKa = 10.12YY340 pKa = 8.58CGKK343 pKa = 8.35MKK345 pKa = 10.48KK346 pKa = 9.67KK347 pKa = 10.83EE348 pKa = 4.04EE349 pKa = 3.99VDD351 pKa = 3.37YY352 pKa = 11.28SLPEE356 pKa = 4.6DD357 pKa = 4.46IIMKK361 pKa = 10.43DD362 pKa = 3.11VDD364 pKa = 3.83EE365 pKa = 5.65DD366 pKa = 4.64SDD368 pKa = 4.66VEE370 pKa = 4.57DD371 pKa = 3.9KK372 pKa = 11.35KK373 pKa = 11.52EE374 pKa = 3.64EE375 pKa = 3.83NAYY378 pKa = 10.74DD379 pKa = 3.75KK380 pKa = 11.42DD381 pKa = 3.34AFRR384 pKa = 11.84PDD386 pKa = 3.17KK387 pKa = 11.19VADD390 pKa = 4.05AGDD393 pKa = 3.89DD394 pKa = 3.8VKK396 pKa = 11.43EE397 pKa = 4.15KK398 pKa = 11.11
MM1 pKa = 7.63EE2 pKa = 6.09KK3 pKa = 9.58LTSLLISCPEE13 pKa = 4.13MEE15 pKa = 5.63QMMQQLLAEE24 pKa = 4.24MQKK27 pKa = 11.14LNATVEE33 pKa = 4.4KK34 pKa = 9.97QNSRR38 pKa = 11.84IEE40 pKa = 3.94QLEE43 pKa = 4.06NEE45 pKa = 4.33LEE47 pKa = 4.11KK48 pKa = 11.24EE49 pKa = 4.07KK50 pKa = 10.96GKK52 pKa = 11.02KK53 pKa = 9.43PLTEE57 pKa = 3.76EE58 pKa = 4.39KK59 pKa = 10.25IISEE63 pKa = 4.21KK64 pKa = 10.56EE65 pKa = 3.76LEE67 pKa = 4.15KK68 pKa = 10.91EE69 pKa = 5.34KK70 pKa = 10.74EE71 pKa = 4.07ISWKK75 pKa = 10.88AKK77 pKa = 7.42IKK79 pKa = 10.54KK80 pKa = 9.85YY81 pKa = 11.03GEE83 pKa = 4.14ASSKK87 pKa = 9.13EE88 pKa = 3.82EE89 pKa = 4.16KK90 pKa = 10.14IIEE93 pKa = 4.06KK94 pKa = 10.55DD95 pKa = 3.04VWQKK99 pKa = 11.21PKK101 pKa = 9.27GTHH104 pKa = 5.61IGSIYY109 pKa = 8.26VTEE112 pKa = 5.2FVRR115 pKa = 11.84LMNYY119 pKa = 10.09LNSRR123 pKa = 11.84HH124 pKa = 6.25ANLNEE129 pKa = 3.96IYY131 pKa = 10.83SLTDD135 pKa = 3.29YY136 pKa = 11.37NKK138 pKa = 10.6LVADD142 pKa = 4.01KK143 pKa = 10.03NTDD146 pKa = 2.97EE147 pKa = 4.2RR148 pKa = 11.84LIRR151 pKa = 11.84AAYY154 pKa = 8.85QHH156 pKa = 6.46GLLHH160 pKa = 4.79VHH162 pKa = 6.58YY163 pKa = 9.9IEE165 pKa = 4.93SANQIKK171 pKa = 9.78MYY173 pKa = 10.87DD174 pKa = 3.45EE175 pKa = 5.58GIVNAYY181 pKa = 10.19LKK183 pKa = 10.57LAQLTKK189 pKa = 10.66AKK191 pKa = 10.0CIYY194 pKa = 9.99IRR196 pKa = 11.84FYY198 pKa = 10.47TAFAEE203 pKa = 4.55VTKK206 pKa = 10.67EE207 pKa = 3.83GIIPKK212 pKa = 9.86IEE214 pKa = 4.5AIKK217 pKa = 10.82LGITYY222 pKa = 9.79EE223 pKa = 4.17KK224 pKa = 10.09IQYY227 pKa = 8.71DD228 pKa = 3.66VCEE231 pKa = 4.03QEE233 pKa = 4.93LFEE236 pKa = 4.82EE237 pKa = 4.34NRR239 pKa = 11.84VHH241 pKa = 7.89DD242 pKa = 4.36FLRR245 pKa = 11.84HH246 pKa = 5.06KK247 pKa = 10.43KK248 pKa = 10.53AIGLLTIRR256 pKa = 11.84RR257 pKa = 11.84EE258 pKa = 4.22LEE260 pKa = 3.95STEE263 pKa = 3.93GNIWVYY269 pKa = 8.98EE270 pKa = 3.9NRR272 pKa = 11.84NNRR275 pKa = 11.84IIYY278 pKa = 9.81APSRR282 pKa = 11.84ARR284 pKa = 11.84KK285 pKa = 9.0EE286 pKa = 3.97EE287 pKa = 3.95VKK289 pKa = 10.66KK290 pKa = 11.08VVAAWQQKK298 pKa = 9.39IYY300 pKa = 9.98TPEE303 pKa = 4.02RR304 pKa = 11.84NIPDD308 pKa = 3.82CGITAPCISEE318 pKa = 4.07PCLKK322 pKa = 10.13ILCEE326 pKa = 3.91LSKK329 pKa = 11.02NQLVVKK335 pKa = 8.42HH336 pKa = 4.65QCKK339 pKa = 10.12YY340 pKa = 8.58CGKK343 pKa = 8.35MKK345 pKa = 10.48KK346 pKa = 9.67KK347 pKa = 10.83EE348 pKa = 4.04EE349 pKa = 3.99VDD351 pKa = 3.37YY352 pKa = 11.28SLPEE356 pKa = 4.6DD357 pKa = 4.46IIMKK361 pKa = 10.43DD362 pKa = 3.11VDD364 pKa = 3.83EE365 pKa = 5.65DD366 pKa = 4.64SDD368 pKa = 4.66VEE370 pKa = 4.57DD371 pKa = 3.9KK372 pKa = 11.35KK373 pKa = 11.52EE374 pKa = 3.64EE375 pKa = 3.83NAYY378 pKa = 10.74DD379 pKa = 3.75KK380 pKa = 11.42DD381 pKa = 3.34AFRR384 pKa = 11.84PDD386 pKa = 3.17KK387 pKa = 11.19VADD390 pKa = 4.05AGDD393 pKa = 3.89DD394 pKa = 3.8VKK396 pKa = 11.43EE397 pKa = 4.15KK398 pKa = 11.11
Molecular weight: 46.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2XXY4|F2XXY4_9VIRU Coat protein/movement protein OS=Sweet potato collusive virus OX=930168 PE=4 SV=1
MM1 pKa = 7.51NRR3 pKa = 11.84MTYY6 pKa = 7.35MTVKK10 pKa = 9.29ITIPTYY16 pKa = 9.17MSRR19 pKa = 11.84IYY21 pKa = 10.52HH22 pKa = 5.97GLFDD26 pKa = 4.17TGANTCVCKK35 pKa = 10.31TKK37 pKa = 10.62VLPPEE42 pKa = 3.61KK43 pKa = 9.59WIEE46 pKa = 3.8IDD48 pKa = 3.66NKK50 pKa = 10.13VLTGFSEE57 pKa = 4.24EE58 pKa = 4.28KK59 pKa = 10.79NVIKK63 pKa = 10.85YY64 pKa = 10.22RR65 pKa = 11.84ADD67 pKa = 3.51NIKK70 pKa = 10.51IMIAKK75 pKa = 9.67KK76 pKa = 10.33EE77 pKa = 4.0FIIPYY82 pKa = 9.06IYY84 pKa = 11.31AMDD87 pKa = 5.36NINADD92 pKa = 3.87IIIGATFYY100 pKa = 11.36NKK102 pKa = 10.1YY103 pKa = 10.71SPITIDD109 pKa = 3.38MEE111 pKa = 4.04KK112 pKa = 10.84GIIKK116 pKa = 8.07FTNKK120 pKa = 10.29GEE122 pKa = 4.17VYY124 pKa = 10.25PNYY127 pKa = 9.91LVKK130 pKa = 10.77YY131 pKa = 8.17PRR133 pKa = 11.84KK134 pKa = 9.44KK135 pKa = 10.86VLIPWEE141 pKa = 4.4KK142 pKa = 10.45GNPSIIQPLQNEE154 pKa = 4.42QINNINEE161 pKa = 3.97VEE163 pKa = 4.16EE164 pKa = 4.25LNNILGEE171 pKa = 4.77DD172 pKa = 3.47IYY174 pKa = 11.56GDD176 pKa = 3.51NPLKK180 pKa = 10.49HH181 pKa = 5.8WEE183 pKa = 3.97KK184 pKa = 10.45HH185 pKa = 3.27KK186 pKa = 9.8TYY188 pKa = 11.25AKK190 pKa = 10.18IEE192 pKa = 4.21LKK194 pKa = 11.05NPDD197 pKa = 3.25DD198 pKa = 5.46HH199 pKa = 8.4IYY201 pKa = 10.46KK202 pKa = 10.44PPINYY207 pKa = 9.16QEE209 pKa = 3.94SDD211 pKa = 3.52YY212 pKa = 11.62KK213 pKa = 9.92EE214 pKa = 4.26FKK216 pKa = 9.41MHH218 pKa = 6.44IDD220 pKa = 3.46EE221 pKa = 4.26MVKK224 pKa = 10.17EE225 pKa = 4.36GFIEE229 pKa = 4.16EE230 pKa = 4.54CKK232 pKa = 10.64NLEE235 pKa = 3.98NKK237 pKa = 10.09KK238 pKa = 9.7YY239 pKa = 10.14SSPAFIVNKK248 pKa = 9.82HH249 pKa = 5.02SEE251 pKa = 3.94IKK253 pKa = 9.97RR254 pKa = 11.84GKK256 pKa = 8.92SRR258 pKa = 11.84MVIDD262 pKa = 4.34YY263 pKa = 10.98KK264 pKa = 11.3DD265 pKa = 3.65LNKK268 pKa = 9.74KK269 pKa = 10.44AKK271 pKa = 9.72VIKK274 pKa = 10.48HH275 pKa = 6.75PIPNKK280 pKa = 10.33DD281 pKa = 2.85ILINRR286 pKa = 11.84GIKK289 pKa = 10.15ANYY292 pKa = 8.88FSKK295 pKa = 10.63FDD297 pKa = 4.02CKK299 pKa = 10.9SGFYY303 pKa = 10.05HH304 pKa = 7.06IKK306 pKa = 10.66LEE308 pKa = 4.19EE309 pKa = 4.4DD310 pKa = 3.45SKK312 pKa = 11.49KK313 pKa = 9.19YY314 pKa = 8.84TAFTVPQGYY323 pKa = 7.93YY324 pKa = 9.0VWIVLPFGYY333 pKa = 10.17HH334 pKa = 6.06NSPSIYY340 pKa = 9.25QQFMDD345 pKa = 5.31GIFRR349 pKa = 11.84PYY351 pKa = 10.14YY352 pKa = 10.54DD353 pKa = 5.22FILVYY358 pKa = 10.17IDD360 pKa = 5.59DD361 pKa = 4.06ILIFSKK367 pKa = 9.69TYY369 pKa = 9.84EE370 pKa = 3.75EE371 pKa = 4.49HH372 pKa = 7.19KK373 pKa = 10.16IHH375 pKa = 7.8LEE377 pKa = 3.42IFRR380 pKa = 11.84NIIIKK385 pKa = 10.35HH386 pKa = 5.96GIVLSKK392 pKa = 10.62KK393 pKa = 9.07KK394 pKa = 10.88AEE396 pKa = 3.95IGKK399 pKa = 9.38QKK401 pKa = 10.55IEE403 pKa = 3.86FLGVKK408 pKa = 9.49IEE410 pKa = 3.93QGGIEE415 pKa = 4.57LQPHH419 pKa = 7.13IIDD422 pKa = 5.09KK423 pKa = 10.69ILEE426 pKa = 3.87KK427 pKa = 10.43HH428 pKa = 6.29IKK430 pKa = 9.63IKK432 pKa = 10.51SKK434 pKa = 11.17KK435 pKa = 8.31EE436 pKa = 3.67LQSILGLVNQIRR448 pKa = 11.84NFLPNLSKK456 pKa = 10.62ILLPIQKK463 pKa = 9.83KK464 pKa = 10.4LKK466 pKa = 10.11IKK468 pKa = 10.81NEE470 pKa = 4.25EE471 pKa = 3.53VWEE474 pKa = 4.11WTKK477 pKa = 11.29EE478 pKa = 3.89DD479 pKa = 3.54EE480 pKa = 4.92QNIIKK485 pKa = 10.71LKK487 pKa = 10.42DD488 pKa = 3.38YY489 pKa = 11.33CKK491 pKa = 11.12DD492 pKa = 3.29NVIKK496 pKa = 8.4MTYY499 pKa = 9.14PVEE502 pKa = 4.18EE503 pKa = 5.14KK504 pKa = 11.01DD505 pKa = 3.58MNWIIEE511 pKa = 3.89VDD513 pKa = 3.37ASKK516 pKa = 10.71EE517 pKa = 3.99YY518 pKa = 11.04YY519 pKa = 10.66GNCLKK524 pKa = 10.84YY525 pKa = 10.56KK526 pKa = 9.6KK527 pKa = 10.4DD528 pKa = 3.42KK529 pKa = 10.8IEE531 pKa = 4.05YY532 pKa = 7.44ICRR535 pKa = 11.84YY536 pKa = 9.69NSGTFKK542 pKa = 10.54EE543 pKa = 4.43HH544 pKa = 6.35EE545 pKa = 4.2KK546 pKa = 11.03NYY548 pKa = 10.52DD549 pKa = 3.16INRR552 pKa = 11.84KK553 pKa = 7.67EE554 pKa = 4.49LIAIYY559 pKa = 10.13KK560 pKa = 9.78GLEE563 pKa = 3.72HH564 pKa = 6.44YY565 pKa = 10.81AIFTTQGKK573 pKa = 9.77KK574 pKa = 9.68LVRR577 pKa = 11.84TDD579 pKa = 3.19NSQAYY584 pKa = 8.67YY585 pKa = 9.54WIKK588 pKa = 10.43NSKK591 pKa = 9.04IKK593 pKa = 10.92NSIDD597 pKa = 3.25MKK599 pKa = 10.15NVKK602 pKa = 10.25GILAKK607 pKa = 10.18IIMYY611 pKa = 10.43DD612 pKa = 3.58FDD614 pKa = 4.77IEE616 pKa = 4.71IIDD619 pKa = 3.88GKK621 pKa = 8.73TNIVADD627 pKa = 4.2FLSRR631 pKa = 11.84NGTDD635 pKa = 3.54DD636 pKa = 3.3ATITGG641 pKa = 3.86
MM1 pKa = 7.51NRR3 pKa = 11.84MTYY6 pKa = 7.35MTVKK10 pKa = 9.29ITIPTYY16 pKa = 9.17MSRR19 pKa = 11.84IYY21 pKa = 10.52HH22 pKa = 5.97GLFDD26 pKa = 4.17TGANTCVCKK35 pKa = 10.31TKK37 pKa = 10.62VLPPEE42 pKa = 3.61KK43 pKa = 9.59WIEE46 pKa = 3.8IDD48 pKa = 3.66NKK50 pKa = 10.13VLTGFSEE57 pKa = 4.24EE58 pKa = 4.28KK59 pKa = 10.79NVIKK63 pKa = 10.85YY64 pKa = 10.22RR65 pKa = 11.84ADD67 pKa = 3.51NIKK70 pKa = 10.51IMIAKK75 pKa = 9.67KK76 pKa = 10.33EE77 pKa = 4.0FIIPYY82 pKa = 9.06IYY84 pKa = 11.31AMDD87 pKa = 5.36NINADD92 pKa = 3.87IIIGATFYY100 pKa = 11.36NKK102 pKa = 10.1YY103 pKa = 10.71SPITIDD109 pKa = 3.38MEE111 pKa = 4.04KK112 pKa = 10.84GIIKK116 pKa = 8.07FTNKK120 pKa = 10.29GEE122 pKa = 4.17VYY124 pKa = 10.25PNYY127 pKa = 9.91LVKK130 pKa = 10.77YY131 pKa = 8.17PRR133 pKa = 11.84KK134 pKa = 9.44KK135 pKa = 10.86VLIPWEE141 pKa = 4.4KK142 pKa = 10.45GNPSIIQPLQNEE154 pKa = 4.42QINNINEE161 pKa = 3.97VEE163 pKa = 4.16EE164 pKa = 4.25LNNILGEE171 pKa = 4.77DD172 pKa = 3.47IYY174 pKa = 11.56GDD176 pKa = 3.51NPLKK180 pKa = 10.49HH181 pKa = 5.8WEE183 pKa = 3.97KK184 pKa = 10.45HH185 pKa = 3.27KK186 pKa = 9.8TYY188 pKa = 11.25AKK190 pKa = 10.18IEE192 pKa = 4.21LKK194 pKa = 11.05NPDD197 pKa = 3.25DD198 pKa = 5.46HH199 pKa = 8.4IYY201 pKa = 10.46KK202 pKa = 10.44PPINYY207 pKa = 9.16QEE209 pKa = 3.94SDD211 pKa = 3.52YY212 pKa = 11.62KK213 pKa = 9.92EE214 pKa = 4.26FKK216 pKa = 9.41MHH218 pKa = 6.44IDD220 pKa = 3.46EE221 pKa = 4.26MVKK224 pKa = 10.17EE225 pKa = 4.36GFIEE229 pKa = 4.16EE230 pKa = 4.54CKK232 pKa = 10.64NLEE235 pKa = 3.98NKK237 pKa = 10.09KK238 pKa = 9.7YY239 pKa = 10.14SSPAFIVNKK248 pKa = 9.82HH249 pKa = 5.02SEE251 pKa = 3.94IKK253 pKa = 9.97RR254 pKa = 11.84GKK256 pKa = 8.92SRR258 pKa = 11.84MVIDD262 pKa = 4.34YY263 pKa = 10.98KK264 pKa = 11.3DD265 pKa = 3.65LNKK268 pKa = 9.74KK269 pKa = 10.44AKK271 pKa = 9.72VIKK274 pKa = 10.48HH275 pKa = 6.75PIPNKK280 pKa = 10.33DD281 pKa = 2.85ILINRR286 pKa = 11.84GIKK289 pKa = 10.15ANYY292 pKa = 8.88FSKK295 pKa = 10.63FDD297 pKa = 4.02CKK299 pKa = 10.9SGFYY303 pKa = 10.05HH304 pKa = 7.06IKK306 pKa = 10.66LEE308 pKa = 4.19EE309 pKa = 4.4DD310 pKa = 3.45SKK312 pKa = 11.49KK313 pKa = 9.19YY314 pKa = 8.84TAFTVPQGYY323 pKa = 7.93YY324 pKa = 9.0VWIVLPFGYY333 pKa = 10.17HH334 pKa = 6.06NSPSIYY340 pKa = 9.25QQFMDD345 pKa = 5.31GIFRR349 pKa = 11.84PYY351 pKa = 10.14YY352 pKa = 10.54DD353 pKa = 5.22FILVYY358 pKa = 10.17IDD360 pKa = 5.59DD361 pKa = 4.06ILIFSKK367 pKa = 9.69TYY369 pKa = 9.84EE370 pKa = 3.75EE371 pKa = 4.49HH372 pKa = 7.19KK373 pKa = 10.16IHH375 pKa = 7.8LEE377 pKa = 3.42IFRR380 pKa = 11.84NIIIKK385 pKa = 10.35HH386 pKa = 5.96GIVLSKK392 pKa = 10.62KK393 pKa = 9.07KK394 pKa = 10.88AEE396 pKa = 3.95IGKK399 pKa = 9.38QKK401 pKa = 10.55IEE403 pKa = 3.86FLGVKK408 pKa = 9.49IEE410 pKa = 3.93QGGIEE415 pKa = 4.57LQPHH419 pKa = 7.13IIDD422 pKa = 5.09KK423 pKa = 10.69ILEE426 pKa = 3.87KK427 pKa = 10.43HH428 pKa = 6.29IKK430 pKa = 9.63IKK432 pKa = 10.51SKK434 pKa = 11.17KK435 pKa = 8.31EE436 pKa = 3.67LQSILGLVNQIRR448 pKa = 11.84NFLPNLSKK456 pKa = 10.62ILLPIQKK463 pKa = 9.83KK464 pKa = 10.4LKK466 pKa = 10.11IKK468 pKa = 10.81NEE470 pKa = 4.25EE471 pKa = 3.53VWEE474 pKa = 4.11WTKK477 pKa = 11.29EE478 pKa = 3.89DD479 pKa = 3.54EE480 pKa = 4.92QNIIKK485 pKa = 10.71LKK487 pKa = 10.42DD488 pKa = 3.38YY489 pKa = 11.33CKK491 pKa = 11.12DD492 pKa = 3.29NVIKK496 pKa = 8.4MTYY499 pKa = 9.14PVEE502 pKa = 4.18EE503 pKa = 5.14KK504 pKa = 11.01DD505 pKa = 3.58MNWIIEE511 pKa = 3.89VDD513 pKa = 3.37ASKK516 pKa = 10.71EE517 pKa = 3.99YY518 pKa = 11.04YY519 pKa = 10.66GNCLKK524 pKa = 10.84YY525 pKa = 10.56KK526 pKa = 9.6KK527 pKa = 10.4DD528 pKa = 3.42KK529 pKa = 10.8IEE531 pKa = 4.05YY532 pKa = 7.44ICRR535 pKa = 11.84YY536 pKa = 9.69NSGTFKK542 pKa = 10.54EE543 pKa = 4.43HH544 pKa = 6.35EE545 pKa = 4.2KK546 pKa = 11.03NYY548 pKa = 10.52DD549 pKa = 3.16INRR552 pKa = 11.84KK553 pKa = 7.67EE554 pKa = 4.49LIAIYY559 pKa = 10.13KK560 pKa = 9.78GLEE563 pKa = 3.72HH564 pKa = 6.44YY565 pKa = 10.81AIFTTQGKK573 pKa = 9.77KK574 pKa = 9.68LVRR577 pKa = 11.84TDD579 pKa = 3.19NSQAYY584 pKa = 8.67YY585 pKa = 9.54WIKK588 pKa = 10.43NSKK591 pKa = 9.04IKK593 pKa = 10.92NSIDD597 pKa = 3.25MKK599 pKa = 10.15NVKK602 pKa = 10.25GILAKK607 pKa = 10.18IIMYY611 pKa = 10.43DD612 pKa = 3.58FDD614 pKa = 4.77IEE616 pKa = 4.71IIDD619 pKa = 3.88GKK621 pKa = 8.73TNIVADD627 pKa = 4.2FLSRR631 pKa = 11.84NGTDD635 pKa = 3.54DD636 pKa = 3.3ATITGG641 pKa = 3.86
Molecular weight: 75.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2352 |
49 |
1264 |
588.0 |
69.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.316 ± 0.986 | 1.19 ± 0.354 |
5.57 ± 0.136 | 10.544 ± 1.23 |
2.976 ± 0.457 | 3.614 ± 0.422 |
1.616 ± 0.435 | 12.287 ± 1.061 |
12.67 ± 0.521 | 6.845 ± 0.475 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.168 ± 0.083 | 7.526 ± 1.045 |
3.316 ± 0.248 | 3.869 ± 0.549 |
3.699 ± 0.593 | 4.677 ± 0.401 |
4.124 ± 0.243 | 3.444 ± 0.777 |
0.978 ± 0.144 | 5.57 ± 0.601 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
