
Kineobactrum salinum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Kineobactrum
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
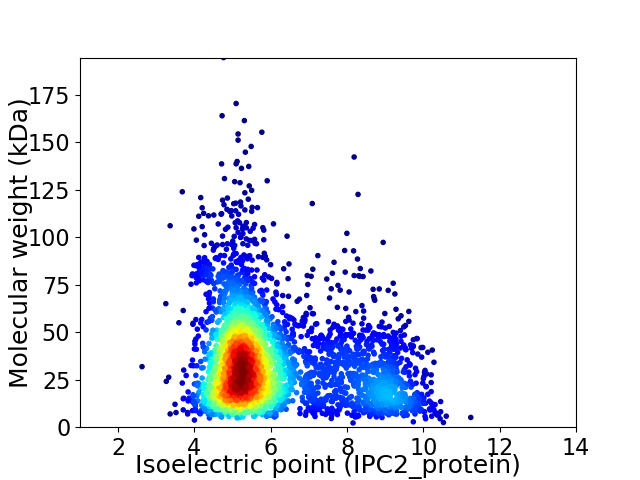
Virtual 2D-PAGE plot for 3712 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6C0U7B6|A0A6C0U7B6_9GAMM Acyl-CoA_dh_1 domain-containing protein OS=Kineobactrum salinum OX=2708301 GN=G3T16_17055 PE=4 SV=1
MM1 pKa = 6.84TCTRR5 pKa = 11.84VSLNAGDD12 pKa = 3.54TWVIGSIGGAVAVSGPSSLSTTASTSSDD40 pKa = 3.41VFDD43 pKa = 5.83GISANNTAAVDD54 pKa = 3.67TTVNEE59 pKa = 4.3GTDD62 pKa = 3.36VSVSMTRR69 pKa = 11.84TGPGTLVQLDD79 pKa = 3.81EE80 pKa = 4.82FVINLNPRR88 pKa = 11.84YY89 pKa = 8.61TGGQPQNLTMTANLDD104 pKa = 3.53PAFQIQNGSPFSQNGWDD121 pKa = 3.76CTVAGQTVNCTRR133 pKa = 11.84ADD135 pKa = 3.31GGGDD139 pKa = 3.11VGGEE143 pKa = 3.74FDD145 pKa = 4.46HH146 pKa = 7.53NIGQVSIDD154 pKa = 3.22VRR156 pKa = 11.84AIAFGNNLGNTATVSSVSPDD176 pKa = 3.49DD177 pKa = 4.05AQLANNSDD185 pKa = 3.75NLNITVQEE193 pKa = 4.17PDD195 pKa = 3.78FDD197 pKa = 4.25LQALKK202 pKa = 10.7SGPTQTLRR210 pKa = 11.84VVNGEE215 pKa = 4.04SFSFNLRR222 pKa = 11.84VRR224 pKa = 11.84NNSNVAFVGEE234 pKa = 4.05LTMTDD239 pKa = 3.63SLPAGLRR246 pKa = 11.84VDD248 pKa = 4.32SYY250 pKa = 12.05SLNGWSCGPATPVNGADD267 pKa = 3.88TISCTRR273 pKa = 11.84TYY275 pKa = 11.09TEE277 pKa = 4.66GSPLAAGATTPSVLLDD293 pKa = 3.72TVATATGSHH302 pKa = 6.23SNQMCVTNTPAPGYY316 pKa = 9.48VFPADD321 pKa = 3.31SDD323 pKa = 4.06GANDD327 pKa = 4.09CDD329 pKa = 3.61NSAVEE334 pKa = 4.3GQLNADD340 pKa = 3.56SADD343 pKa = 3.22IQVYY347 pKa = 9.95KK348 pKa = 10.56SGSATVVAGEE358 pKa = 3.99LLSYY362 pKa = 10.46QLEE365 pKa = 4.36IVNAGPEE372 pKa = 4.13PAQSVTLTDD381 pKa = 3.39TFGSLFTGGPGNGFEE396 pKa = 4.36GAVVVAGNATLGSCSNAAAANVTSRR421 pKa = 11.84NLSCTFSDD429 pKa = 4.72MPVCSQGVDD438 pKa = 3.49CPVITVQVRR447 pKa = 11.84PLGSASNGTDD457 pKa = 3.21LNRR460 pKa = 11.84NNTASAFSSVTSDD473 pKa = 3.08PVYY476 pKa = 11.22GNNSDD481 pKa = 3.47NHH483 pKa = 5.62ATTVTGQADD492 pKa = 3.02IAAIKK497 pKa = 7.56TASWAEE503 pKa = 4.06DD504 pKa = 3.98PDD506 pKa = 5.23PIAAGTNLSYY516 pKa = 10.78TITVRR521 pKa = 11.84NEE523 pKa = 3.49STGASGASTVSMSDD537 pKa = 3.37TLPLDD542 pKa = 3.28VTFISASASDD552 pKa = 3.91GGSCGVTPAVGATTEE567 pKa = 4.1AGDD570 pKa = 3.73RR571 pKa = 11.84TVEE574 pKa = 4.3CTWASVQRR582 pKa = 11.84GGQRR586 pKa = 11.84TVSLVVRR593 pKa = 11.84PNRR596 pKa = 11.84GTQASTLSNTVTSATSTPEE615 pKa = 3.57VTLDD619 pKa = 3.79NNDD622 pKa = 3.55ADD624 pKa = 3.68VDD626 pKa = 4.37VPVAAPEE633 pKa = 3.91VDD635 pKa = 3.59LATTQVDD642 pKa = 3.98SPDD645 pKa = 3.66PLQIDD650 pKa = 3.66EE651 pKa = 4.68TVVYY655 pKa = 9.38TLTVRR660 pKa = 11.84NLGPSVAEE668 pKa = 3.83TVRR671 pKa = 11.84LYY673 pKa = 11.16NVLPTSGLSFLGFQYY688 pKa = 11.11VDD690 pKa = 3.42GGGITQPGLPPGVSCPVQPAVGEE713 pKa = 4.04FGKK716 pKa = 8.75TVDD719 pKa = 4.33RR720 pKa = 11.84VDD722 pKa = 3.55TALLTPSDD730 pKa = 4.16PAWLNGAWDD739 pKa = 4.22PEE741 pKa = 4.44LDD743 pKa = 3.91NADD746 pKa = 4.32IICDD750 pKa = 3.6VSALMLAGDD759 pKa = 5.0DD760 pKa = 4.65ISFQMLLDD768 pKa = 4.16AEE770 pKa = 4.4ASGVYY775 pKa = 8.26TNYY778 pKa = 10.17MIARR782 pKa = 11.84SQEE785 pKa = 3.81HH786 pKa = 7.06RR787 pKa = 11.84DD788 pKa = 3.47GFDD791 pKa = 3.69DD792 pKa = 4.37VNPANDD798 pKa = 4.56LEE800 pKa = 4.54PEE802 pKa = 4.07NTTVRR807 pKa = 11.84TRR809 pKa = 11.84ADD811 pKa = 3.25MEE813 pKa = 4.62VASKK817 pKa = 8.99TADD820 pKa = 3.64PGSVSLFEE828 pKa = 4.23PFEE831 pKa = 4.03YY832 pKa = 10.64SIVVQNNGPHH842 pKa = 6.14EE843 pKa = 4.31ALGVEE848 pKa = 4.73LEE850 pKa = 4.48DD851 pKa = 3.84SFPAGMEE858 pKa = 4.14LLGAPAVVVDD868 pKa = 3.68QGSFEE873 pKa = 4.31TTSCSGVAGDD883 pKa = 4.37TAVTCDD889 pKa = 3.99FGEE892 pKa = 4.16ISADD896 pKa = 2.98GRR898 pKa = 11.84ATVTLPVRR906 pKa = 11.84LISAAGAAVNNTATVNVTGLTLDD929 pKa = 3.99DD930 pKa = 3.91TPGNNSNDD938 pKa = 3.0GSVTVVRR945 pKa = 11.84SSIAGRR951 pKa = 11.84VYY953 pKa = 10.45HH954 pKa = 6.8DD955 pKa = 3.48QDD957 pKa = 2.79ISGDD961 pKa = 3.81YY962 pKa = 10.93DD963 pKa = 3.45VGEE966 pKa = 4.29PGISGVTVTLTGSDD980 pKa = 3.73DD981 pKa = 3.59YY982 pKa = 11.8GNTVNRR988 pKa = 11.84STTTDD993 pKa = 2.73GTGEE997 pKa = 3.87YY998 pKa = 9.84RR999 pKa = 11.84FEE1001 pKa = 4.95DD1002 pKa = 4.23LAPGVYY1008 pKa = 9.42TLTEE1012 pKa = 4.0SHH1014 pKa = 6.67PAAWVDD1020 pKa = 3.59GQEE1023 pKa = 4.51TVGSVGGTTPTTDD1036 pKa = 2.86VITNIEE1042 pKa = 3.92LAAGVDD1048 pKa = 3.54ATDD1051 pKa = 3.4YY1052 pKa = 11.28NYY1054 pKa = 11.42GEE1056 pKa = 4.24FQVGSVASISGYY1068 pKa = 10.3VYY1070 pKa = 10.43HH1071 pKa = 7.32DD1072 pKa = 3.68QNEE1075 pKa = 4.11DD1076 pKa = 3.92GIFDD1080 pKa = 3.89GDD1082 pKa = 3.9EE1083 pKa = 3.85NPIAGVVVDD1092 pKa = 4.81LAGPVPGSTTTDD1104 pKa = 2.87ANGFYY1109 pKa = 10.29SFTGLAPGSYY1119 pKa = 8.78TVTEE1123 pKa = 4.03QQPTDD1128 pKa = 3.41WLDD1131 pKa = 3.84GLDD1134 pKa = 3.67TAGTVAALVSEE1145 pKa = 4.36VDD1147 pKa = 3.63DD1148 pKa = 4.31EE1149 pKa = 5.09FVVEE1153 pKa = 5.13LLAGDD1158 pKa = 5.23EE1159 pKa = 4.07AQQWNFGEE1167 pKa = 4.29IALPDD1172 pKa = 3.65APAPVPALNRR1182 pKa = 11.84TALALLSALLLLLSAGHH1199 pKa = 6.07LRR1201 pKa = 11.84RR1202 pKa = 11.84RR1203 pKa = 11.84GLLL1206 pKa = 3.44
MM1 pKa = 6.84TCTRR5 pKa = 11.84VSLNAGDD12 pKa = 3.54TWVIGSIGGAVAVSGPSSLSTTASTSSDD40 pKa = 3.41VFDD43 pKa = 5.83GISANNTAAVDD54 pKa = 3.67TTVNEE59 pKa = 4.3GTDD62 pKa = 3.36VSVSMTRR69 pKa = 11.84TGPGTLVQLDD79 pKa = 3.81EE80 pKa = 4.82FVINLNPRR88 pKa = 11.84YY89 pKa = 8.61TGGQPQNLTMTANLDD104 pKa = 3.53PAFQIQNGSPFSQNGWDD121 pKa = 3.76CTVAGQTVNCTRR133 pKa = 11.84ADD135 pKa = 3.31GGGDD139 pKa = 3.11VGGEE143 pKa = 3.74FDD145 pKa = 4.46HH146 pKa = 7.53NIGQVSIDD154 pKa = 3.22VRR156 pKa = 11.84AIAFGNNLGNTATVSSVSPDD176 pKa = 3.49DD177 pKa = 4.05AQLANNSDD185 pKa = 3.75NLNITVQEE193 pKa = 4.17PDD195 pKa = 3.78FDD197 pKa = 4.25LQALKK202 pKa = 10.7SGPTQTLRR210 pKa = 11.84VVNGEE215 pKa = 4.04SFSFNLRR222 pKa = 11.84VRR224 pKa = 11.84NNSNVAFVGEE234 pKa = 4.05LTMTDD239 pKa = 3.63SLPAGLRR246 pKa = 11.84VDD248 pKa = 4.32SYY250 pKa = 12.05SLNGWSCGPATPVNGADD267 pKa = 3.88TISCTRR273 pKa = 11.84TYY275 pKa = 11.09TEE277 pKa = 4.66GSPLAAGATTPSVLLDD293 pKa = 3.72TVATATGSHH302 pKa = 6.23SNQMCVTNTPAPGYY316 pKa = 9.48VFPADD321 pKa = 3.31SDD323 pKa = 4.06GANDD327 pKa = 4.09CDD329 pKa = 3.61NSAVEE334 pKa = 4.3GQLNADD340 pKa = 3.56SADD343 pKa = 3.22IQVYY347 pKa = 9.95KK348 pKa = 10.56SGSATVVAGEE358 pKa = 3.99LLSYY362 pKa = 10.46QLEE365 pKa = 4.36IVNAGPEE372 pKa = 4.13PAQSVTLTDD381 pKa = 3.39TFGSLFTGGPGNGFEE396 pKa = 4.36GAVVVAGNATLGSCSNAAAANVTSRR421 pKa = 11.84NLSCTFSDD429 pKa = 4.72MPVCSQGVDD438 pKa = 3.49CPVITVQVRR447 pKa = 11.84PLGSASNGTDD457 pKa = 3.21LNRR460 pKa = 11.84NNTASAFSSVTSDD473 pKa = 3.08PVYY476 pKa = 11.22GNNSDD481 pKa = 3.47NHH483 pKa = 5.62ATTVTGQADD492 pKa = 3.02IAAIKK497 pKa = 7.56TASWAEE503 pKa = 4.06DD504 pKa = 3.98PDD506 pKa = 5.23PIAAGTNLSYY516 pKa = 10.78TITVRR521 pKa = 11.84NEE523 pKa = 3.49STGASGASTVSMSDD537 pKa = 3.37TLPLDD542 pKa = 3.28VTFISASASDD552 pKa = 3.91GGSCGVTPAVGATTEE567 pKa = 4.1AGDD570 pKa = 3.73RR571 pKa = 11.84TVEE574 pKa = 4.3CTWASVQRR582 pKa = 11.84GGQRR586 pKa = 11.84TVSLVVRR593 pKa = 11.84PNRR596 pKa = 11.84GTQASTLSNTVTSATSTPEE615 pKa = 3.57VTLDD619 pKa = 3.79NNDD622 pKa = 3.55ADD624 pKa = 3.68VDD626 pKa = 4.37VPVAAPEE633 pKa = 3.91VDD635 pKa = 3.59LATTQVDD642 pKa = 3.98SPDD645 pKa = 3.66PLQIDD650 pKa = 3.66EE651 pKa = 4.68TVVYY655 pKa = 9.38TLTVRR660 pKa = 11.84NLGPSVAEE668 pKa = 3.83TVRR671 pKa = 11.84LYY673 pKa = 11.16NVLPTSGLSFLGFQYY688 pKa = 11.11VDD690 pKa = 3.42GGGITQPGLPPGVSCPVQPAVGEE713 pKa = 4.04FGKK716 pKa = 8.75TVDD719 pKa = 4.33RR720 pKa = 11.84VDD722 pKa = 3.55TALLTPSDD730 pKa = 4.16PAWLNGAWDD739 pKa = 4.22PEE741 pKa = 4.44LDD743 pKa = 3.91NADD746 pKa = 4.32IICDD750 pKa = 3.6VSALMLAGDD759 pKa = 5.0DD760 pKa = 4.65ISFQMLLDD768 pKa = 4.16AEE770 pKa = 4.4ASGVYY775 pKa = 8.26TNYY778 pKa = 10.17MIARR782 pKa = 11.84SQEE785 pKa = 3.81HH786 pKa = 7.06RR787 pKa = 11.84DD788 pKa = 3.47GFDD791 pKa = 3.69DD792 pKa = 4.37VNPANDD798 pKa = 4.56LEE800 pKa = 4.54PEE802 pKa = 4.07NTTVRR807 pKa = 11.84TRR809 pKa = 11.84ADD811 pKa = 3.25MEE813 pKa = 4.62VASKK817 pKa = 8.99TADD820 pKa = 3.64PGSVSLFEE828 pKa = 4.23PFEE831 pKa = 4.03YY832 pKa = 10.64SIVVQNNGPHH842 pKa = 6.14EE843 pKa = 4.31ALGVEE848 pKa = 4.73LEE850 pKa = 4.48DD851 pKa = 3.84SFPAGMEE858 pKa = 4.14LLGAPAVVVDD868 pKa = 3.68QGSFEE873 pKa = 4.31TTSCSGVAGDD883 pKa = 4.37TAVTCDD889 pKa = 3.99FGEE892 pKa = 4.16ISADD896 pKa = 2.98GRR898 pKa = 11.84ATVTLPVRR906 pKa = 11.84LISAAGAAVNNTATVNVTGLTLDD929 pKa = 3.99DD930 pKa = 3.91TPGNNSNDD938 pKa = 3.0GSVTVVRR945 pKa = 11.84SSIAGRR951 pKa = 11.84VYY953 pKa = 10.45HH954 pKa = 6.8DD955 pKa = 3.48QDD957 pKa = 2.79ISGDD961 pKa = 3.81YY962 pKa = 10.93DD963 pKa = 3.45VGEE966 pKa = 4.29PGISGVTVTLTGSDD980 pKa = 3.73DD981 pKa = 3.59YY982 pKa = 11.8GNTVNRR988 pKa = 11.84STTTDD993 pKa = 2.73GTGEE997 pKa = 3.87YY998 pKa = 9.84RR999 pKa = 11.84FEE1001 pKa = 4.95DD1002 pKa = 4.23LAPGVYY1008 pKa = 9.42TLTEE1012 pKa = 4.0SHH1014 pKa = 6.67PAAWVDD1020 pKa = 3.59GQEE1023 pKa = 4.51TVGSVGGTTPTTDD1036 pKa = 2.86VITNIEE1042 pKa = 3.92LAAGVDD1048 pKa = 3.54ATDD1051 pKa = 3.4YY1052 pKa = 11.28NYY1054 pKa = 11.42GEE1056 pKa = 4.24FQVGSVASISGYY1068 pKa = 10.3VYY1070 pKa = 10.43HH1071 pKa = 7.32DD1072 pKa = 3.68QNEE1075 pKa = 4.11DD1076 pKa = 3.92GIFDD1080 pKa = 3.89GDD1082 pKa = 3.9EE1083 pKa = 3.85NPIAGVVVDD1092 pKa = 4.81LAGPVPGSTTTDD1104 pKa = 2.87ANGFYY1109 pKa = 10.29SFTGLAPGSYY1119 pKa = 8.78TVTEE1123 pKa = 4.03QQPTDD1128 pKa = 3.41WLDD1131 pKa = 3.84GLDD1134 pKa = 3.67TAGTVAALVSEE1145 pKa = 4.36VDD1147 pKa = 3.63DD1148 pKa = 4.31EE1149 pKa = 5.09FVVEE1153 pKa = 5.13LLAGDD1158 pKa = 5.23EE1159 pKa = 4.07AQQWNFGEE1167 pKa = 4.29IALPDD1172 pKa = 3.65APAPVPALNRR1182 pKa = 11.84TALALLSALLLLLSAGHH1199 pKa = 6.07LRR1201 pKa = 11.84RR1202 pKa = 11.84RR1203 pKa = 11.84GLLL1206 pKa = 3.44
Molecular weight: 124.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6C0U0I5|A0A6C0U0I5_9GAMM Phosphopantetheine adenylyltransferase OS=Kineobactrum salinum OX=2708301 GN=coaD PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.42RR12 pKa = 11.84ARR14 pKa = 11.84AHH16 pKa = 6.29GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.42RR12 pKa = 11.84ARR14 pKa = 11.84AHH16 pKa = 6.29GFRR19 pKa = 11.84SRR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.99GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1225903 |
20 |
1787 |
330.3 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.009 ± 0.052 | 1.063 ± 0.013 |
5.746 ± 0.031 | 6.162 ± 0.035 |
3.556 ± 0.027 | 8.139 ± 0.037 |
2.225 ± 0.02 | 4.802 ± 0.032 |
2.665 ± 0.032 | 11.053 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.262 ± 0.019 | 2.934 ± 0.023 |
4.899 ± 0.03 | 4.225 ± 0.031 |
7.05 ± 0.041 | 5.942 ± 0.03 |
5.049 ± 0.028 | 7.104 ± 0.033 |
1.406 ± 0.017 | 2.708 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
