
Thalassobius mediterraneus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Thalassobius
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
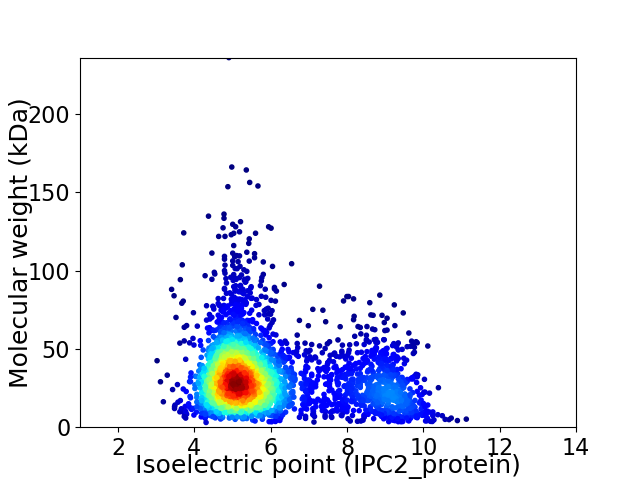
Virtual 2D-PAGE plot for 3260 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P1GLN9|A0A0P1GLN9_9RHOB Glycerol-3-phosphate acyltransferase OS=Thalassobius mediterraneus OX=340021 GN=plsB PE=4 SV=1
MM1 pKa = 7.6PGNRR5 pKa = 11.84HH6 pKa = 5.04HH7 pKa = 7.17HH8 pKa = 6.06LFGLQGQLQGQGQMQGQGQHH28 pKa = 5.44QGQGQHH34 pKa = 5.56QGQGQGQHH42 pKa = 5.42QGQHH46 pKa = 5.45SEE48 pKa = 3.88NLNLNGNGNMNGNGNLNGNGNHH70 pKa = 6.68NGNEE74 pKa = 3.99NGNGNEE80 pKa = 4.1NHH82 pKa = 5.67NTNHH86 pKa = 5.69NTTSSDD92 pKa = 3.81AISAGLGMGVGAGLGHH108 pKa = 6.96AHH110 pKa = 7.27AGANSDD116 pKa = 3.97SGSHH120 pKa = 6.04SGSEE124 pKa = 4.18ASSSASTSNHH134 pKa = 4.97TDD136 pKa = 2.68VGVSVAVDD144 pKa = 3.62LTDD147 pKa = 4.34WEE149 pKa = 4.35PTDD152 pKa = 5.83DD153 pKa = 5.52DD154 pKa = 4.62FADD157 pKa = 4.13FDD159 pKa = 4.21LHH161 pKa = 6.94NSAVGDD167 pKa = 3.37ITMADD172 pKa = 3.23GDD174 pKa = 4.52MNTDD178 pKa = 2.47IGGNMSLDD186 pKa = 3.4GALNNALDD194 pKa = 4.09GAGNDD199 pKa = 3.46TGVINAMSNSLQDD212 pKa = 3.47NDD214 pKa = 3.86NLEE217 pKa = 4.45GASVSNGGSYY227 pKa = 8.66TATMTAHH234 pKa = 6.99GGSADD239 pKa = 3.72SEE241 pKa = 4.08EE242 pKa = 5.51GIDD245 pKa = 3.79NEE247 pKa = 4.76ADD249 pKa = 3.33GGSAGSAYY257 pKa = 10.69SDD259 pKa = 3.42GGKK262 pKa = 10.71GSTGNANGGHH272 pKa = 6.4SIALGGKK279 pKa = 9.5HH280 pKa = 5.81YY281 pKa = 11.14GNDD284 pKa = 3.67GGKK287 pKa = 9.52SEE289 pKa = 4.96GDD291 pKa = 3.6DD292 pKa = 4.37GGDD295 pKa = 3.42SGNAGAIAVGLGGLGLGLGGSADD318 pKa = 3.69GGTTGAGGAGGAGGTGGAGGSGGNGGNAGSAGGIGAAQGGDD359 pKa = 3.44SGALGAGAGGDD370 pKa = 3.5AGAIGAGAGGDD381 pKa = 3.53AGAVGAGLGGGLGLLPIGTGMGVGDD406 pKa = 4.33ADD408 pKa = 3.78AGNGNGTAGATAGAGTGTAGANAGNAGGYY437 pKa = 10.35GLGNSQSGGNDD448 pKa = 3.17ADD450 pKa = 4.19GAAGGAGAAGGAGAGSGTADD470 pKa = 2.93GGMAGGMGMGMGMGGGTAGAATGAGGAGGAGGDD503 pKa = 3.56AGASGGVAGGAVGYY517 pKa = 10.04GGHH520 pKa = 5.38ATIWGGNGGASGGSSADD537 pKa = 3.33GGAGGQTGGSWGSYY551 pKa = 9.88NGDD554 pKa = 3.31DD555 pKa = 4.02GFVNGYY561 pKa = 7.31STSSADD567 pKa = 3.71AALDD571 pKa = 3.43NAAFNQNIVQGANVLGNTVDD591 pKa = 3.48MTVVGGGYY599 pKa = 10.0SSTLIGDD606 pKa = 4.41DD607 pKa = 4.21DD608 pKa = 5.08LSS610 pKa = 3.89
MM1 pKa = 7.6PGNRR5 pKa = 11.84HH6 pKa = 5.04HH7 pKa = 7.17HH8 pKa = 6.06LFGLQGQLQGQGQMQGQGQHH28 pKa = 5.44QGQGQHH34 pKa = 5.56QGQGQGQHH42 pKa = 5.42QGQHH46 pKa = 5.45SEE48 pKa = 3.88NLNLNGNGNMNGNGNLNGNGNHH70 pKa = 6.68NGNEE74 pKa = 3.99NGNGNEE80 pKa = 4.1NHH82 pKa = 5.67NTNHH86 pKa = 5.69NTTSSDD92 pKa = 3.81AISAGLGMGVGAGLGHH108 pKa = 6.96AHH110 pKa = 7.27AGANSDD116 pKa = 3.97SGSHH120 pKa = 6.04SGSEE124 pKa = 4.18ASSSASTSNHH134 pKa = 4.97TDD136 pKa = 2.68VGVSVAVDD144 pKa = 3.62LTDD147 pKa = 4.34WEE149 pKa = 4.35PTDD152 pKa = 5.83DD153 pKa = 5.52DD154 pKa = 4.62FADD157 pKa = 4.13FDD159 pKa = 4.21LHH161 pKa = 6.94NSAVGDD167 pKa = 3.37ITMADD172 pKa = 3.23GDD174 pKa = 4.52MNTDD178 pKa = 2.47IGGNMSLDD186 pKa = 3.4GALNNALDD194 pKa = 4.09GAGNDD199 pKa = 3.46TGVINAMSNSLQDD212 pKa = 3.47NDD214 pKa = 3.86NLEE217 pKa = 4.45GASVSNGGSYY227 pKa = 8.66TATMTAHH234 pKa = 6.99GGSADD239 pKa = 3.72SEE241 pKa = 4.08EE242 pKa = 5.51GIDD245 pKa = 3.79NEE247 pKa = 4.76ADD249 pKa = 3.33GGSAGSAYY257 pKa = 10.69SDD259 pKa = 3.42GGKK262 pKa = 10.71GSTGNANGGHH272 pKa = 6.4SIALGGKK279 pKa = 9.5HH280 pKa = 5.81YY281 pKa = 11.14GNDD284 pKa = 3.67GGKK287 pKa = 9.52SEE289 pKa = 4.96GDD291 pKa = 3.6DD292 pKa = 4.37GGDD295 pKa = 3.42SGNAGAIAVGLGGLGLGLGGSADD318 pKa = 3.69GGTTGAGGAGGAGGTGGAGGSGGNGGNAGSAGGIGAAQGGDD359 pKa = 3.44SGALGAGAGGDD370 pKa = 3.5AGAIGAGAGGDD381 pKa = 3.53AGAVGAGLGGGLGLLPIGTGMGVGDD406 pKa = 4.33ADD408 pKa = 3.78AGNGNGTAGATAGAGTGTAGANAGNAGGYY437 pKa = 10.35GLGNSQSGGNDD448 pKa = 3.17ADD450 pKa = 4.19GAAGGAGAAGGAGAGSGTADD470 pKa = 2.93GGMAGGMGMGMGMGGGTAGAATGAGGAGGAGGDD503 pKa = 3.56AGASGGVAGGAVGYY517 pKa = 10.04GGHH520 pKa = 5.38ATIWGGNGGASGGSSADD537 pKa = 3.33GGAGGQTGGSWGSYY551 pKa = 9.88NGDD554 pKa = 3.31DD555 pKa = 4.02GFVNGYY561 pKa = 7.31STSSADD567 pKa = 3.71AALDD571 pKa = 3.43NAAFNQNIVQGANVLGNTVDD591 pKa = 3.48MTVVGGGYY599 pKa = 10.0SSTLIGDD606 pKa = 4.41DD607 pKa = 4.21DD608 pKa = 5.08LSS610 pKa = 3.89
Molecular weight: 55.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P1GPW6|A0A0P1GPW6_9RHOB Phospho-2-dehydro-3-deoxyheptonate aldolase OS=Thalassobius mediterraneus OX=340021 GN=aroG PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1013264 |
29 |
2169 |
310.8 |
33.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.146 ± 0.054 | 0.912 ± 0.014 |
6.227 ± 0.038 | 5.896 ± 0.042 |
3.657 ± 0.027 | 8.441 ± 0.047 |
2.115 ± 0.023 | 5.099 ± 0.029 |
3.471 ± 0.04 | 10.117 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.923 ± 0.021 | 2.788 ± 0.022 |
4.805 ± 0.031 | 3.679 ± 0.024 |
6.278 ± 0.042 | 5.112 ± 0.027 |
5.458 ± 0.028 | 7.27 ± 0.038 |
1.33 ± 0.019 | 2.275 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
