
Sphingomonas solaris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
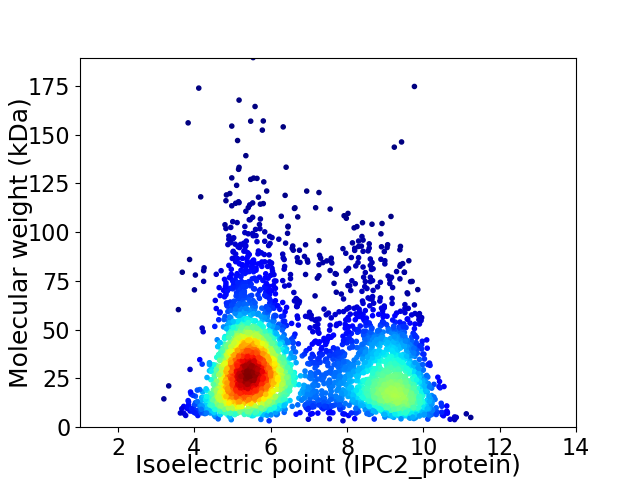
Virtual 2D-PAGE plot for 4080 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A558RB38|A0A558RB38_9SPHN MFS transporter OS=Sphingomonas solaris OX=2529389 GN=FOY91_03600 PE=4 SV=1
VV1 pKa = 4.67VTGTGEE7 pKa = 3.74AGATVVISTADD18 pKa = 3.63GVVLGSKK25 pKa = 10.13VVGSDD30 pKa = 2.96GTYY33 pKa = 10.84LVVLDD38 pKa = 3.77PAQRR42 pKa = 11.84NGEE45 pKa = 4.17ALVATQRR52 pKa = 11.84DD53 pKa = 3.68PAGNVSDD60 pKa = 4.43PVGLSAPDD68 pKa = 3.46LTAPPPPGTLAVDD81 pKa = 3.98DD82 pKa = 5.68DD83 pKa = 4.3GTTLTGAGEE92 pKa = 3.87AGARR96 pKa = 11.84VEE98 pKa = 4.88VRR100 pKa = 11.84DD101 pKa = 4.14PDD103 pKa = 3.78GTLLGSGTVAPDD115 pKa = 3.48GSFTVEE121 pKa = 4.18LAPAQTAGEE130 pKa = 4.29TLMVTQADD138 pKa = 3.57AAGNVSGPASIDD150 pKa = 3.33APFDD154 pKa = 3.3VQAYY158 pKa = 10.37DD159 pKa = 3.45NFRR162 pKa = 11.84TASIDD167 pKa = 3.71LEE169 pKa = 4.35PTTTSVDD176 pKa = 3.37HH177 pKa = 7.01GSANYY182 pKa = 9.43LALVSLGLVNLQAEE196 pKa = 4.66VLSVNNVRR204 pKa = 11.84FSVQAGHH211 pKa = 6.56SLDD214 pKa = 5.38AEE216 pKa = 4.25FTYY219 pKa = 11.1NALLNIGVASGYY231 pKa = 9.79SVVVQRR237 pKa = 11.84LEE239 pKa = 3.98GTQWVAVSGTGPASVLEE256 pKa = 4.18LGLLNGDD263 pKa = 3.93LRR265 pKa = 11.84ATEE268 pKa = 4.1TFGPGEE274 pKa = 3.89YY275 pKa = 9.51RR276 pKa = 11.84AFLTFDD282 pKa = 3.49GAAGVGLLGNLNVTGSEE299 pKa = 4.08ADD301 pKa = 3.61FTANADD307 pKa = 3.62VTPIAATGNVIRR319 pKa = 11.84DD320 pKa = 3.78AGPDD324 pKa = 3.7GNVDD328 pKa = 3.19AVTPGTLLTSVTVNGVPIPVLPGGSTINTVWGTLTIAPDD367 pKa = 3.58GNYY370 pKa = 10.33SYY372 pKa = 11.3LPNADD377 pKa = 3.42PAAIGRR383 pKa = 11.84TEE385 pKa = 4.01VFTYY389 pKa = 10.55TLFDD393 pKa = 4.27PSSGQIEE400 pKa = 4.71SATLSITIDD409 pKa = 3.56SPDD412 pKa = 3.2IAGPPVAMPDD422 pKa = 3.43TAVAAVTFEE431 pKa = 4.18NVVAIAPSAPTFGFTSQPSGLLQSTGSGNGTFTVAANAEE470 pKa = 3.96ADD472 pKa = 3.4ITITAVRR479 pKa = 11.84QAGLSVSLLPTYY491 pKa = 9.95TVTLHH496 pKa = 6.33NADD499 pKa = 3.43NSVVRR504 pKa = 11.84TEE506 pKa = 4.28TVTALASALLGTAASFTFNDD526 pKa = 4.29LPGGAYY532 pKa = 8.87TYY534 pKa = 9.16TVSSSATTLGNFGTTVSLGSATTFLDD560 pKa = 4.16TYY562 pKa = 11.55ALATRR567 pKa = 11.84EE568 pKa = 4.33TVTGSLVDD576 pKa = 3.56NDD578 pKa = 3.98TTNTPFTTIRR588 pKa = 11.84INAGAGMVEE597 pKa = 4.29IDD599 pKa = 3.47QAPVSFAGQYY609 pKa = 7.52GTLTVDD615 pKa = 3.15QTGHH619 pKa = 4.73YY620 pKa = 8.36TYY622 pKa = 10.59QPHH625 pKa = 5.62EE626 pKa = 4.16TLAYY630 pKa = 10.16SATDD634 pKa = 3.41LVDD637 pKa = 3.09TFTYY641 pKa = 10.81QFVQPNGVVSVSTLNVTIDD660 pKa = 3.5VPGDD664 pKa = 3.52GPALVATTTSLAMAPSGHH682 pKa = 6.51EE683 pKa = 3.93GDD685 pKa = 5.25VIPLDD690 pKa = 4.08ALTHH694 pKa = 5.46QPGGDD699 pKa = 3.06AAVAQDD705 pKa = 3.74VAVSLATYY713 pKa = 10.73DD714 pKa = 3.71LFEE717 pKa = 4.58GQGDD721 pKa = 4.22LEE723 pKa = 4.85DD724 pKa = 3.73VLSNYY729 pKa = 10.19LSARR733 pKa = 11.84HH734 pKa = 6.59DD735 pKa = 3.82GATVVADD742 pKa = 3.65TASYY746 pKa = 10.68SSNSIDD752 pKa = 3.98LSGSTVTDD760 pKa = 3.55PFDD763 pKa = 3.72YY764 pKa = 10.88LVTVNDD770 pKa = 3.95HH771 pKa = 6.59DD772 pKa = 4.22HH773 pKa = 7.02DD774 pKa = 4.11RR775 pKa = 11.84TVSQHH780 pKa = 5.6VMM782 pKa = 3.19
VV1 pKa = 4.67VTGTGEE7 pKa = 3.74AGATVVISTADD18 pKa = 3.63GVVLGSKK25 pKa = 10.13VVGSDD30 pKa = 2.96GTYY33 pKa = 10.84LVVLDD38 pKa = 3.77PAQRR42 pKa = 11.84NGEE45 pKa = 4.17ALVATQRR52 pKa = 11.84DD53 pKa = 3.68PAGNVSDD60 pKa = 4.43PVGLSAPDD68 pKa = 3.46LTAPPPPGTLAVDD81 pKa = 3.98DD82 pKa = 5.68DD83 pKa = 4.3GTTLTGAGEE92 pKa = 3.87AGARR96 pKa = 11.84VEE98 pKa = 4.88VRR100 pKa = 11.84DD101 pKa = 4.14PDD103 pKa = 3.78GTLLGSGTVAPDD115 pKa = 3.48GSFTVEE121 pKa = 4.18LAPAQTAGEE130 pKa = 4.29TLMVTQADD138 pKa = 3.57AAGNVSGPASIDD150 pKa = 3.33APFDD154 pKa = 3.3VQAYY158 pKa = 10.37DD159 pKa = 3.45NFRR162 pKa = 11.84TASIDD167 pKa = 3.71LEE169 pKa = 4.35PTTTSVDD176 pKa = 3.37HH177 pKa = 7.01GSANYY182 pKa = 9.43LALVSLGLVNLQAEE196 pKa = 4.66VLSVNNVRR204 pKa = 11.84FSVQAGHH211 pKa = 6.56SLDD214 pKa = 5.38AEE216 pKa = 4.25FTYY219 pKa = 11.1NALLNIGVASGYY231 pKa = 9.79SVVVQRR237 pKa = 11.84LEE239 pKa = 3.98GTQWVAVSGTGPASVLEE256 pKa = 4.18LGLLNGDD263 pKa = 3.93LRR265 pKa = 11.84ATEE268 pKa = 4.1TFGPGEE274 pKa = 3.89YY275 pKa = 9.51RR276 pKa = 11.84AFLTFDD282 pKa = 3.49GAAGVGLLGNLNVTGSEE299 pKa = 4.08ADD301 pKa = 3.61FTANADD307 pKa = 3.62VTPIAATGNVIRR319 pKa = 11.84DD320 pKa = 3.78AGPDD324 pKa = 3.7GNVDD328 pKa = 3.19AVTPGTLLTSVTVNGVPIPVLPGGSTINTVWGTLTIAPDD367 pKa = 3.58GNYY370 pKa = 10.33SYY372 pKa = 11.3LPNADD377 pKa = 3.42PAAIGRR383 pKa = 11.84TEE385 pKa = 4.01VFTYY389 pKa = 10.55TLFDD393 pKa = 4.27PSSGQIEE400 pKa = 4.71SATLSITIDD409 pKa = 3.56SPDD412 pKa = 3.2IAGPPVAMPDD422 pKa = 3.43TAVAAVTFEE431 pKa = 4.18NVVAIAPSAPTFGFTSQPSGLLQSTGSGNGTFTVAANAEE470 pKa = 3.96ADD472 pKa = 3.4ITITAVRR479 pKa = 11.84QAGLSVSLLPTYY491 pKa = 9.95TVTLHH496 pKa = 6.33NADD499 pKa = 3.43NSVVRR504 pKa = 11.84TEE506 pKa = 4.28TVTALASALLGTAASFTFNDD526 pKa = 4.29LPGGAYY532 pKa = 8.87TYY534 pKa = 9.16TVSSSATTLGNFGTTVSLGSATTFLDD560 pKa = 4.16TYY562 pKa = 11.55ALATRR567 pKa = 11.84EE568 pKa = 4.33TVTGSLVDD576 pKa = 3.56NDD578 pKa = 3.98TTNTPFTTIRR588 pKa = 11.84INAGAGMVEE597 pKa = 4.29IDD599 pKa = 3.47QAPVSFAGQYY609 pKa = 7.52GTLTVDD615 pKa = 3.15QTGHH619 pKa = 4.73YY620 pKa = 8.36TYY622 pKa = 10.59QPHH625 pKa = 5.62EE626 pKa = 4.16TLAYY630 pKa = 10.16SATDD634 pKa = 3.41LVDD637 pKa = 3.09TFTYY641 pKa = 10.81QFVQPNGVVSVSTLNVTIDD660 pKa = 3.5VPGDD664 pKa = 3.52GPALVATTTSLAMAPSGHH682 pKa = 6.51EE683 pKa = 3.93GDD685 pKa = 5.25VIPLDD690 pKa = 4.08ALTHH694 pKa = 5.46QPGGDD699 pKa = 3.06AAVAQDD705 pKa = 3.74VAVSLATYY713 pKa = 10.73DD714 pKa = 3.71LFEE717 pKa = 4.58GQGDD721 pKa = 4.22LEE723 pKa = 4.85DD724 pKa = 3.73VLSNYY729 pKa = 10.19LSARR733 pKa = 11.84HH734 pKa = 6.59DD735 pKa = 3.82GATVVADD742 pKa = 3.65TASYY746 pKa = 10.68SSNSIDD752 pKa = 3.98LSGSTVTDD760 pKa = 3.55PFDD763 pKa = 3.72YY764 pKa = 10.88LVTVNDD770 pKa = 3.95HH771 pKa = 6.59DD772 pKa = 4.22HH773 pKa = 7.02DD774 pKa = 4.11RR775 pKa = 11.84TVSQHH780 pKa = 5.6VMM782 pKa = 3.19
Molecular weight: 79.44 kDa
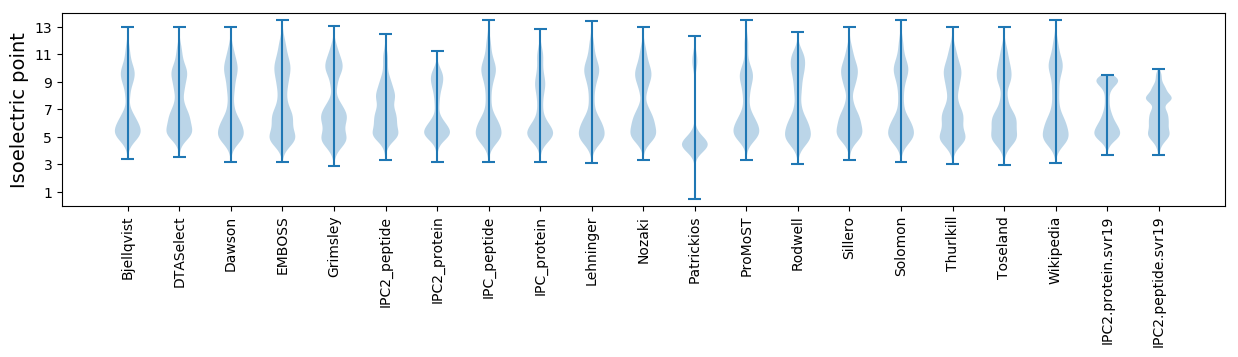
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A558R008|A0A558R008_9SPHN 50S ribosomal protein L16 OS=Sphingomonas solaris OX=2529389 GN=rplP PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.32GFRR19 pKa = 11.84TRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 8.76ILAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84IKK41 pKa = 10.86LSAA44 pKa = 3.84
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.32GFRR19 pKa = 11.84TRR21 pKa = 11.84SATPGGRR28 pKa = 11.84KK29 pKa = 8.76ILAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84IKK41 pKa = 10.86LSAA44 pKa = 3.84
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1288240 |
29 |
1886 |
315.7 |
33.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.312 ± 0.062 | 0.763 ± 0.012 |
6.074 ± 0.028 | 4.984 ± 0.037 |
3.37 ± 0.021 | 9.25 ± 0.04 |
1.972 ± 0.021 | 4.773 ± 0.023 |
2.53 ± 0.026 | 9.896 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.298 ± 0.02 | 2.271 ± 0.026 |
5.561 ± 0.03 | 2.771 ± 0.021 |
7.919 ± 0.041 | 4.836 ± 0.03 |
5.529 ± 0.034 | 7.407 ± 0.027 |
1.365 ± 0.017 | 2.121 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
