
Butyrivibrio sp. TB
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio; unclassified Butyrivibrio
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
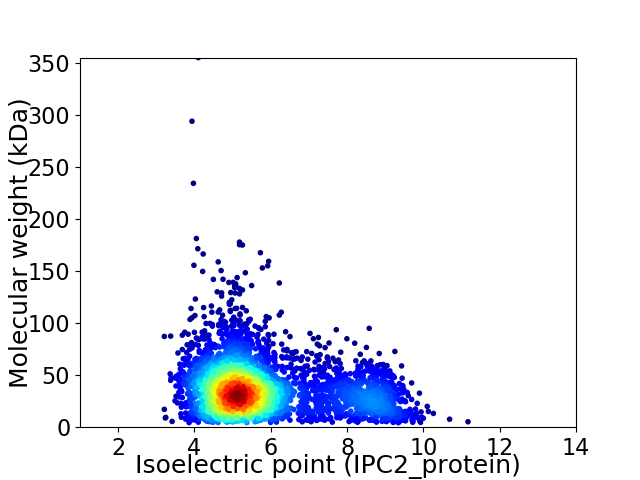
Virtual 2D-PAGE plot for 3714 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
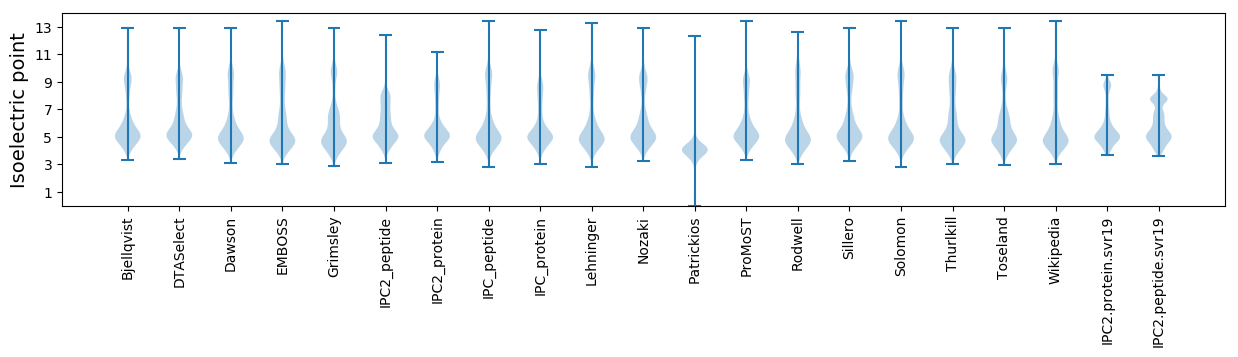
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9GRL2|A0A1H9GRL2_9FIRM Non-specific serine/threonine protein kinase OS=Butyrivibrio sp. TB OX=1520809 GN=SAMN02910382_03317 PE=4 SV=1
MM1 pKa = 7.78KK2 pKa = 10.29KK3 pKa = 10.36RR4 pKa = 11.84IISVMLGAMILCAGCGAAINIEE26 pKa = 3.98EE27 pKa = 4.69DD28 pKa = 3.87EE29 pKa = 4.35ITSEE33 pKa = 4.18DD34 pKa = 3.55KK35 pKa = 11.44VEE37 pKa = 3.98EE38 pKa = 4.17FEE40 pKa = 4.37DD41 pKa = 3.83AKK43 pKa = 11.1SEE45 pKa = 4.17SSNEE49 pKa = 3.7VATISSEE56 pKa = 3.93EE57 pKa = 4.23SSVDD61 pKa = 3.08TSEE64 pKa = 5.28EE65 pKa = 3.45IDD67 pKa = 3.41ITDD70 pKa = 4.57EE71 pKa = 4.54EE72 pKa = 5.71DD73 pKa = 3.52DD74 pKa = 3.95TTKK77 pKa = 10.59DD78 pKa = 2.92TDD80 pKa = 3.45TSKK83 pKa = 11.59DD84 pKa = 3.52EE85 pKa = 5.07DD86 pKa = 4.45DD87 pKa = 3.84STNDD91 pKa = 3.0VSRR94 pKa = 11.84YY95 pKa = 8.57EE96 pKa = 4.27SFVSNTEE103 pKa = 3.65EE104 pKa = 4.22VFFDD108 pKa = 3.82EE109 pKa = 4.89SGSDD113 pKa = 3.21ILPYY117 pKa = 10.39SLQRR121 pKa = 11.84NFDD124 pKa = 3.44LSKK127 pKa = 10.83GYY129 pKa = 8.93TIDD132 pKa = 4.22EE133 pKa = 4.19IVEE136 pKa = 4.17ASLDD140 pKa = 3.9SYY142 pKa = 11.21TSYY145 pKa = 11.52DD146 pKa = 3.85DD147 pKa = 5.39SKK149 pKa = 11.5DD150 pKa = 3.65DD151 pKa = 4.05PSFEE155 pKa = 4.33TKK157 pKa = 10.87YY158 pKa = 10.37IDD160 pKa = 4.82CGNDD164 pKa = 3.94GIDD167 pKa = 3.84EE168 pKa = 4.58LLVDD172 pKa = 3.62IRR174 pKa = 11.84FPGMEE179 pKa = 3.92TADD182 pKa = 3.47YY183 pKa = 10.55TDD185 pKa = 3.88YY186 pKa = 11.36EE187 pKa = 4.39EE188 pKa = 6.06KK189 pKa = 10.96YY190 pKa = 9.49VDD192 pKa = 2.68RR193 pKa = 11.84WVIKK197 pKa = 10.51EE198 pKa = 4.08IDD200 pKa = 3.71DD201 pKa = 3.89KK202 pKa = 11.87LIVKK206 pKa = 9.2YY207 pKa = 10.75VGALRR212 pKa = 11.84HH213 pKa = 5.6EE214 pKa = 5.09SYY216 pKa = 10.67TIINDD221 pKa = 3.06YY222 pKa = 11.06GYY224 pKa = 10.5IKK226 pKa = 10.6NEE228 pKa = 3.49VMGAYY233 pKa = 10.12NGVLSEE239 pKa = 4.35SYY241 pKa = 10.97GYY243 pKa = 11.2LDD245 pKa = 5.67ADD247 pKa = 4.51ANWVLYY253 pKa = 10.04YY254 pKa = 10.58KK255 pKa = 10.71YY256 pKa = 10.85NYY258 pKa = 9.42YY259 pKa = 10.92LDD261 pKa = 3.49MDD263 pKa = 4.6DD264 pKa = 4.1YY265 pKa = 11.84VRR267 pKa = 11.84EE268 pKa = 4.15KK269 pKa = 11.31GNYY272 pKa = 9.96GYY274 pKa = 10.88DD275 pKa = 3.9LDD277 pKa = 4.8CSSDD281 pKa = 2.87GWSNMQVEE289 pKa = 5.12EE290 pKa = 4.92YY291 pKa = 10.51IFDD294 pKa = 4.38DD295 pKa = 3.94EE296 pKa = 5.72ARR298 pKa = 11.84ITDD301 pKa = 3.76SKK303 pKa = 11.64DD304 pKa = 3.01SGFITYY310 pKa = 9.92YY311 pKa = 10.61KK312 pKa = 9.89YY313 pKa = 11.07DD314 pKa = 3.35SDD316 pKa = 5.26GRR318 pKa = 11.84IEE320 pKa = 5.0DD321 pKa = 3.61DD322 pKa = 4.87SIYY325 pKa = 10.5EE326 pKa = 4.15DD327 pKa = 3.18SSIYY331 pKa = 10.61KK332 pKa = 8.71KK333 pKa = 9.3TFDD336 pKa = 4.13DD337 pKa = 3.86VDD339 pKa = 4.28VNICPYY345 pKa = 10.68SEE347 pKa = 4.0VEE349 pKa = 3.83KK350 pKa = 10.68RR351 pKa = 11.84LEE353 pKa = 3.85EE354 pKa = 3.89RR355 pKa = 11.84RR356 pKa = 11.84EE357 pKa = 3.96EE358 pKa = 3.97IGLSEE363 pKa = 4.81DD364 pKa = 3.75VMGG367 pKa = 5.24
MM1 pKa = 7.78KK2 pKa = 10.29KK3 pKa = 10.36RR4 pKa = 11.84IISVMLGAMILCAGCGAAINIEE26 pKa = 3.98EE27 pKa = 4.69DD28 pKa = 3.87EE29 pKa = 4.35ITSEE33 pKa = 4.18DD34 pKa = 3.55KK35 pKa = 11.44VEE37 pKa = 3.98EE38 pKa = 4.17FEE40 pKa = 4.37DD41 pKa = 3.83AKK43 pKa = 11.1SEE45 pKa = 4.17SSNEE49 pKa = 3.7VATISSEE56 pKa = 3.93EE57 pKa = 4.23SSVDD61 pKa = 3.08TSEE64 pKa = 5.28EE65 pKa = 3.45IDD67 pKa = 3.41ITDD70 pKa = 4.57EE71 pKa = 4.54EE72 pKa = 5.71DD73 pKa = 3.52DD74 pKa = 3.95TTKK77 pKa = 10.59DD78 pKa = 2.92TDD80 pKa = 3.45TSKK83 pKa = 11.59DD84 pKa = 3.52EE85 pKa = 5.07DD86 pKa = 4.45DD87 pKa = 3.84STNDD91 pKa = 3.0VSRR94 pKa = 11.84YY95 pKa = 8.57EE96 pKa = 4.27SFVSNTEE103 pKa = 3.65EE104 pKa = 4.22VFFDD108 pKa = 3.82EE109 pKa = 4.89SGSDD113 pKa = 3.21ILPYY117 pKa = 10.39SLQRR121 pKa = 11.84NFDD124 pKa = 3.44LSKK127 pKa = 10.83GYY129 pKa = 8.93TIDD132 pKa = 4.22EE133 pKa = 4.19IVEE136 pKa = 4.17ASLDD140 pKa = 3.9SYY142 pKa = 11.21TSYY145 pKa = 11.52DD146 pKa = 3.85DD147 pKa = 5.39SKK149 pKa = 11.5DD150 pKa = 3.65DD151 pKa = 4.05PSFEE155 pKa = 4.33TKK157 pKa = 10.87YY158 pKa = 10.37IDD160 pKa = 4.82CGNDD164 pKa = 3.94GIDD167 pKa = 3.84EE168 pKa = 4.58LLVDD172 pKa = 3.62IRR174 pKa = 11.84FPGMEE179 pKa = 3.92TADD182 pKa = 3.47YY183 pKa = 10.55TDD185 pKa = 3.88YY186 pKa = 11.36EE187 pKa = 4.39EE188 pKa = 6.06KK189 pKa = 10.96YY190 pKa = 9.49VDD192 pKa = 2.68RR193 pKa = 11.84WVIKK197 pKa = 10.51EE198 pKa = 4.08IDD200 pKa = 3.71DD201 pKa = 3.89KK202 pKa = 11.87LIVKK206 pKa = 9.2YY207 pKa = 10.75VGALRR212 pKa = 11.84HH213 pKa = 5.6EE214 pKa = 5.09SYY216 pKa = 10.67TIINDD221 pKa = 3.06YY222 pKa = 11.06GYY224 pKa = 10.5IKK226 pKa = 10.6NEE228 pKa = 3.49VMGAYY233 pKa = 10.12NGVLSEE239 pKa = 4.35SYY241 pKa = 10.97GYY243 pKa = 11.2LDD245 pKa = 5.67ADD247 pKa = 4.51ANWVLYY253 pKa = 10.04YY254 pKa = 10.58KK255 pKa = 10.71YY256 pKa = 10.85NYY258 pKa = 9.42YY259 pKa = 10.92LDD261 pKa = 3.49MDD263 pKa = 4.6DD264 pKa = 4.1YY265 pKa = 11.84VRR267 pKa = 11.84EE268 pKa = 4.15KK269 pKa = 11.31GNYY272 pKa = 9.96GYY274 pKa = 10.88DD275 pKa = 3.9LDD277 pKa = 4.8CSSDD281 pKa = 2.87GWSNMQVEE289 pKa = 5.12EE290 pKa = 4.92YY291 pKa = 10.51IFDD294 pKa = 4.38DD295 pKa = 3.94EE296 pKa = 5.72ARR298 pKa = 11.84ITDD301 pKa = 3.76SKK303 pKa = 11.64DD304 pKa = 3.01SGFITYY310 pKa = 9.92YY311 pKa = 10.61KK312 pKa = 9.89YY313 pKa = 11.07DD314 pKa = 3.35SDD316 pKa = 5.26GRR318 pKa = 11.84IEE320 pKa = 5.0DD321 pKa = 3.61DD322 pKa = 4.87SIYY325 pKa = 10.5EE326 pKa = 4.15DD327 pKa = 3.18SSIYY331 pKa = 10.61KK332 pKa = 8.71KK333 pKa = 9.3TFDD336 pKa = 4.13DD337 pKa = 3.86VDD339 pKa = 4.28VNICPYY345 pKa = 10.68SEE347 pKa = 4.0VEE349 pKa = 3.83KK350 pKa = 10.68RR351 pKa = 11.84LEE353 pKa = 3.85EE354 pKa = 3.89RR355 pKa = 11.84RR356 pKa = 11.84EE357 pKa = 3.96EE358 pKa = 3.97IGLSEE363 pKa = 4.81DD364 pKa = 3.75VMGG367 pKa = 5.24
Molecular weight: 42.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9GMT9|A0A1H9GMT9_9FIRM SpoIIIJ-associated protein OS=Butyrivibrio sp. TB OX=1520809 GN=SAMN02910382_03271 PE=4 SV=1
MM1 pKa = 7.7AKK3 pKa = 8.06MTFQPHH9 pKa = 5.63KK10 pKa = 8.81LQRR13 pKa = 11.84KK14 pKa = 7.68RR15 pKa = 11.84VHH17 pKa = 6.29GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTPGGRR29 pKa = 11.84KK30 pKa = 8.7VLAARR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.38GRR40 pKa = 11.84KK41 pKa = 8.94RR42 pKa = 11.84LSAA45 pKa = 3.96
MM1 pKa = 7.7AKK3 pKa = 8.06MTFQPHH9 pKa = 5.63KK10 pKa = 8.81LQRR13 pKa = 11.84KK14 pKa = 7.68RR15 pKa = 11.84VHH17 pKa = 6.29GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTPGGRR29 pKa = 11.84KK30 pKa = 8.7VLAARR35 pKa = 11.84RR36 pKa = 11.84LKK38 pKa = 10.38GRR40 pKa = 11.84KK41 pKa = 8.94RR42 pKa = 11.84LSAA45 pKa = 3.96
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1257469 |
39 |
3300 |
338.6 |
38.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.028 ± 0.035 | 1.375 ± 0.015 |
6.931 ± 0.039 | 6.915 ± 0.042 |
4.28 ± 0.028 | 6.826 ± 0.036 |
1.671 ± 0.016 | 7.988 ± 0.04 |
6.808 ± 0.036 | 8.398 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.024 ± 0.023 | 4.732 ± 0.031 |
3.063 ± 0.02 | 2.739 ± 0.021 |
3.999 ± 0.029 | 6.619 ± 0.045 |
5.457 ± 0.038 | 6.6 ± 0.031 |
0.909 ± 0.014 | 4.639 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
