
Tortoise microvirus 88
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.79
Get precalculated fractions of proteins
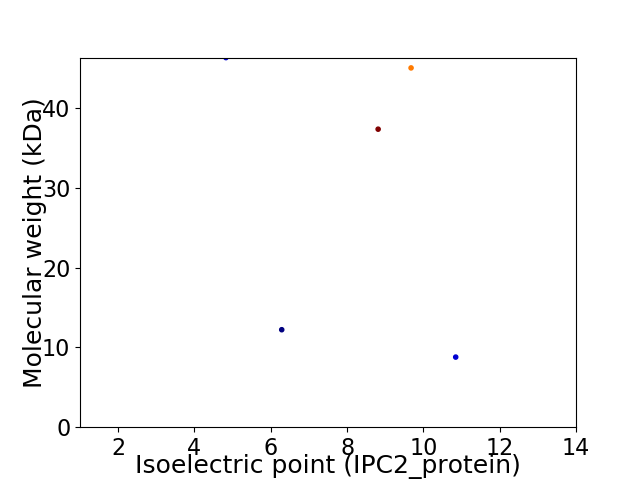
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8WAA5|A0A4P8WAA5_9VIRU Major capsid protein OS=Tortoise microvirus 88 OX=2583196 PE=4 SV=1
MM1 pKa = 7.29ARR3 pKa = 11.84SVTVRR8 pKa = 11.84QLAQSVKK15 pKa = 9.77PMGRR19 pKa = 11.84VMRR22 pKa = 11.84RR23 pKa = 11.84PQHH26 pKa = 5.09TFNIRR31 pKa = 11.84HH32 pKa = 6.23RR33 pKa = 11.84PWQIQPFMIAPVLPGEE49 pKa = 4.29TMKK52 pKa = 11.16NLLLQSRR59 pKa = 11.84VVSDD63 pKa = 3.87PVKK66 pKa = 10.65HH67 pKa = 6.28PLIGWWQEE75 pKa = 3.67YY76 pKa = 8.57YY77 pKa = 10.76FFYY80 pKa = 10.85VKK82 pKa = 10.47HH83 pKa = 6.87RR84 pKa = 11.84DD85 pKa = 3.23LEE87 pKa = 4.35GRR89 pKa = 11.84DD90 pKa = 4.69DD91 pKa = 4.05FTQMMLDD98 pKa = 3.84PQHH101 pKa = 6.49EE102 pKa = 4.61LSSLDD107 pKa = 3.53TPANALTYY115 pKa = 10.43HH116 pKa = 6.97GLTGLDD122 pKa = 3.27WTAMCLEE129 pKa = 4.21RR130 pKa = 11.84VVAEE134 pKa = 3.93YY135 pKa = 10.57FRR137 pKa = 11.84DD138 pKa = 3.95EE139 pKa = 4.16EE140 pKa = 4.33DD141 pKa = 3.34LASVFEE147 pKa = 4.75LGGLPLASISRR158 pKa = 11.84SDD160 pKa = 3.59FAQSLVTATEE170 pKa = 4.44FAARR174 pKa = 11.84QPEE177 pKa = 4.46DD178 pKa = 3.53APADD182 pKa = 3.75ADD184 pKa = 3.54ASGVITASEE193 pKa = 4.11IEE195 pKa = 4.56GALQMWQFQRR205 pKa = 11.84SNGLTKK211 pKa = 9.48MDD213 pKa = 4.25YY214 pKa = 11.13EE215 pKa = 5.01DD216 pKa = 5.16FLATYY221 pKa = 8.79GVNMPKK227 pKa = 10.46EE228 pKa = 4.17EE229 pKa = 4.03LHH231 pKa = 6.14KK232 pKa = 10.74PEE234 pKa = 5.51LIRR237 pKa = 11.84YY238 pKa = 7.71VRR240 pKa = 11.84DD241 pKa = 2.86WTYY244 pKa = 10.62PANTVDD250 pKa = 4.44PSTGTPSSALSWAVAEE266 pKa = 4.68SADD269 pKa = 3.29KK270 pKa = 11.19DD271 pKa = 3.73RR272 pKa = 11.84FFRR275 pKa = 11.84EE276 pKa = 3.6PGFIFGVSVCRR287 pKa = 11.84PKK289 pKa = 10.76VYY291 pKa = 10.6YY292 pKa = 10.43SRR294 pKa = 11.84QTSAAVSMMTDD305 pKa = 3.2AFSWLPAVTSDD316 pKa = 5.69DD317 pKa = 3.49PWTSLKK323 pKa = 10.34PFQAGEE329 pKa = 4.65GPFSGATEE337 pKa = 4.77GYY339 pKa = 9.4WIDD342 pKa = 3.87IKK344 pKa = 11.38DD345 pKa = 3.71LLLYY349 pKa = 10.62GDD351 pKa = 3.64QFINFVPAVGEE362 pKa = 4.07AGFVALPTAGLQRR375 pKa = 11.84KK376 pKa = 7.75FASDD380 pKa = 3.29ADD382 pKa = 3.99ANALFTDD389 pKa = 3.81VAKK392 pKa = 11.03NLIRR396 pKa = 11.84SDD398 pKa = 4.25GIVSLGIAGQQVDD411 pKa = 4.13TTPVV415 pKa = 2.79
MM1 pKa = 7.29ARR3 pKa = 11.84SVTVRR8 pKa = 11.84QLAQSVKK15 pKa = 9.77PMGRR19 pKa = 11.84VMRR22 pKa = 11.84RR23 pKa = 11.84PQHH26 pKa = 5.09TFNIRR31 pKa = 11.84HH32 pKa = 6.23RR33 pKa = 11.84PWQIQPFMIAPVLPGEE49 pKa = 4.29TMKK52 pKa = 11.16NLLLQSRR59 pKa = 11.84VVSDD63 pKa = 3.87PVKK66 pKa = 10.65HH67 pKa = 6.28PLIGWWQEE75 pKa = 3.67YY76 pKa = 8.57YY77 pKa = 10.76FFYY80 pKa = 10.85VKK82 pKa = 10.47HH83 pKa = 6.87RR84 pKa = 11.84DD85 pKa = 3.23LEE87 pKa = 4.35GRR89 pKa = 11.84DD90 pKa = 4.69DD91 pKa = 4.05FTQMMLDD98 pKa = 3.84PQHH101 pKa = 6.49EE102 pKa = 4.61LSSLDD107 pKa = 3.53TPANALTYY115 pKa = 10.43HH116 pKa = 6.97GLTGLDD122 pKa = 3.27WTAMCLEE129 pKa = 4.21RR130 pKa = 11.84VVAEE134 pKa = 3.93YY135 pKa = 10.57FRR137 pKa = 11.84DD138 pKa = 3.95EE139 pKa = 4.16EE140 pKa = 4.33DD141 pKa = 3.34LASVFEE147 pKa = 4.75LGGLPLASISRR158 pKa = 11.84SDD160 pKa = 3.59FAQSLVTATEE170 pKa = 4.44FAARR174 pKa = 11.84QPEE177 pKa = 4.46DD178 pKa = 3.53APADD182 pKa = 3.75ADD184 pKa = 3.54ASGVITASEE193 pKa = 4.11IEE195 pKa = 4.56GALQMWQFQRR205 pKa = 11.84SNGLTKK211 pKa = 9.48MDD213 pKa = 4.25YY214 pKa = 11.13EE215 pKa = 5.01DD216 pKa = 5.16FLATYY221 pKa = 8.79GVNMPKK227 pKa = 10.46EE228 pKa = 4.17EE229 pKa = 4.03LHH231 pKa = 6.14KK232 pKa = 10.74PEE234 pKa = 5.51LIRR237 pKa = 11.84YY238 pKa = 7.71VRR240 pKa = 11.84DD241 pKa = 2.86WTYY244 pKa = 10.62PANTVDD250 pKa = 4.44PSTGTPSSALSWAVAEE266 pKa = 4.68SADD269 pKa = 3.29KK270 pKa = 11.19DD271 pKa = 3.73RR272 pKa = 11.84FFRR275 pKa = 11.84EE276 pKa = 3.6PGFIFGVSVCRR287 pKa = 11.84PKK289 pKa = 10.76VYY291 pKa = 10.6YY292 pKa = 10.43SRR294 pKa = 11.84QTSAAVSMMTDD305 pKa = 3.2AFSWLPAVTSDD316 pKa = 5.69DD317 pKa = 3.49PWTSLKK323 pKa = 10.34PFQAGEE329 pKa = 4.65GPFSGATEE337 pKa = 4.77GYY339 pKa = 9.4WIDD342 pKa = 3.87IKK344 pKa = 11.38DD345 pKa = 3.71LLLYY349 pKa = 10.62GDD351 pKa = 3.64QFINFVPAVGEE362 pKa = 4.07AGFVALPTAGLQRR375 pKa = 11.84KK376 pKa = 7.75FASDD380 pKa = 3.29ADD382 pKa = 3.99ANALFTDD389 pKa = 3.81VAKK392 pKa = 11.03NLIRR396 pKa = 11.84SDD398 pKa = 4.25GIVSLGIAGQQVDD411 pKa = 4.13TTPVV415 pKa = 2.79
Molecular weight: 46.29 kDa
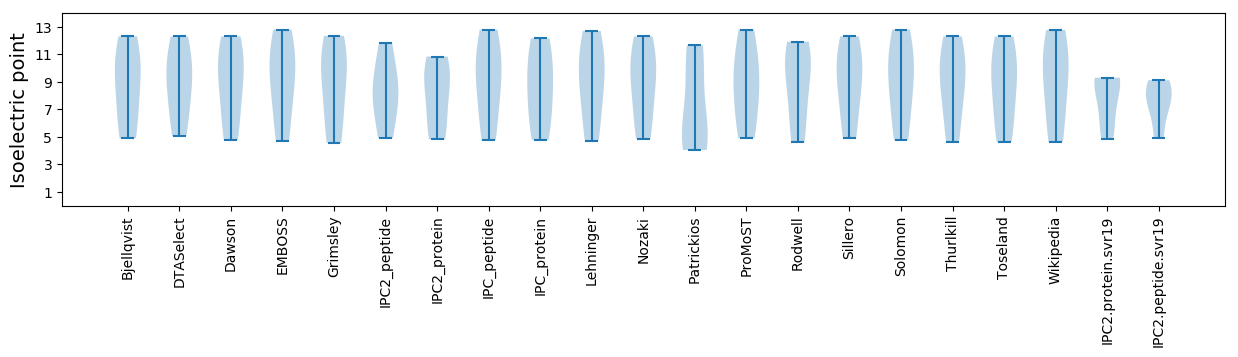
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8WA53|A0A4P8WA53_9VIRU Replication initiation protein OS=Tortoise microvirus 88 OX=2583196 PE=4 SV=1
MM1 pKa = 7.27NMSALAKK8 pKa = 9.3GYY10 pKa = 9.47PMPSPVRR17 pKa = 11.84PQGMTGYY24 pKa = 9.75QRR26 pKa = 11.84QPLRR30 pKa = 11.84LPVPANDD37 pKa = 3.77NPRR40 pKa = 11.84PRR42 pKa = 11.84PRR44 pKa = 11.84PANDD48 pKa = 3.01NFRR51 pKa = 11.84PTPRR55 pKa = 11.84LPPAKK60 pKa = 9.18PFSPPRR66 pKa = 11.84VAPKK70 pKa = 10.41GAWLGYY76 pKa = 9.97KK77 pKa = 10.11LAARR81 pKa = 11.84GAARR85 pKa = 11.84FIPYY89 pKa = 10.26LGAALTAFEE98 pKa = 4.54IYY100 pKa = 9.36EE101 pKa = 3.74WWRR104 pKa = 11.84AGEE107 pKa = 4.18VVPEE111 pKa = 4.26VPEE114 pKa = 4.75HH115 pKa = 6.09IALNGWRR122 pKa = 11.84HH123 pKa = 5.73VGHH126 pKa = 6.71CPKK129 pKa = 10.88GGGGWTHH136 pKa = 7.17RR137 pKa = 11.84GWGDD141 pKa = 3.44QPPTQTQINQANSCLPGQGLSVWLPWGSPYY171 pKa = 10.02PANSRR176 pKa = 11.84TLIAFKK182 pKa = 8.99EE183 pKa = 4.19THH185 pKa = 7.03RR186 pKa = 11.84IGTTPYY192 pKa = 9.68GEE194 pKa = 4.02VRR196 pKa = 11.84EE197 pKa = 4.37VFSRR201 pKa = 11.84PVGGTVTWPEE211 pKa = 3.98YY212 pKa = 10.59VPYY215 pKa = 10.44SPAVLPDD222 pKa = 3.46VVPSPRR228 pKa = 11.84SVPEE232 pKa = 3.45VHH234 pKa = 6.93PGIDD238 pKa = 3.65PFSVPPLAPVAPPAPLPVRR257 pKa = 11.84ALPYY261 pKa = 9.95RR262 pKa = 11.84VSNPMRR268 pKa = 11.84AEE270 pKa = 3.6QSVRR274 pKa = 11.84GFGVNPRR281 pKa = 11.84SVPRR285 pKa = 11.84VNPRR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84PPGVRR296 pKa = 11.84EE297 pKa = 3.95RR298 pKa = 11.84KK299 pKa = 9.35FKK301 pKa = 10.28LTISGSIVSNAFSFVTEE318 pKa = 4.1GADD321 pKa = 3.3VVEE324 pKa = 5.61AIYY327 pKa = 10.14EE328 pKa = 4.52AIPEE332 pKa = 4.52SIRR335 pKa = 11.84GTRR338 pKa = 11.84KK339 pKa = 10.1RR340 pKa = 11.84GVKK343 pKa = 8.7EE344 pKa = 3.15QAYY347 pKa = 8.6FIWQHH352 pKa = 5.98LSQVDD357 pKa = 3.8QNVMVQNLLYY367 pKa = 11.0NHH369 pKa = 6.71FEE371 pKa = 4.43DD372 pKa = 5.09KK373 pKa = 10.89FFGAIGEE380 pKa = 4.26RR381 pKa = 11.84MKK383 pKa = 10.74RR384 pKa = 11.84ANQVVWEE391 pKa = 4.04QGYY394 pKa = 11.02GNLRR398 pKa = 11.84VQFGLAHH405 pKa = 6.79
MM1 pKa = 7.27NMSALAKK8 pKa = 9.3GYY10 pKa = 9.47PMPSPVRR17 pKa = 11.84PQGMTGYY24 pKa = 9.75QRR26 pKa = 11.84QPLRR30 pKa = 11.84LPVPANDD37 pKa = 3.77NPRR40 pKa = 11.84PRR42 pKa = 11.84PRR44 pKa = 11.84PANDD48 pKa = 3.01NFRR51 pKa = 11.84PTPRR55 pKa = 11.84LPPAKK60 pKa = 9.18PFSPPRR66 pKa = 11.84VAPKK70 pKa = 10.41GAWLGYY76 pKa = 9.97KK77 pKa = 10.11LAARR81 pKa = 11.84GAARR85 pKa = 11.84FIPYY89 pKa = 10.26LGAALTAFEE98 pKa = 4.54IYY100 pKa = 9.36EE101 pKa = 3.74WWRR104 pKa = 11.84AGEE107 pKa = 4.18VVPEE111 pKa = 4.26VPEE114 pKa = 4.75HH115 pKa = 6.09IALNGWRR122 pKa = 11.84HH123 pKa = 5.73VGHH126 pKa = 6.71CPKK129 pKa = 10.88GGGGWTHH136 pKa = 7.17RR137 pKa = 11.84GWGDD141 pKa = 3.44QPPTQTQINQANSCLPGQGLSVWLPWGSPYY171 pKa = 10.02PANSRR176 pKa = 11.84TLIAFKK182 pKa = 8.99EE183 pKa = 4.19THH185 pKa = 7.03RR186 pKa = 11.84IGTTPYY192 pKa = 9.68GEE194 pKa = 4.02VRR196 pKa = 11.84EE197 pKa = 4.37VFSRR201 pKa = 11.84PVGGTVTWPEE211 pKa = 3.98YY212 pKa = 10.59VPYY215 pKa = 10.44SPAVLPDD222 pKa = 3.46VVPSPRR228 pKa = 11.84SVPEE232 pKa = 3.45VHH234 pKa = 6.93PGIDD238 pKa = 3.65PFSVPPLAPVAPPAPLPVRR257 pKa = 11.84ALPYY261 pKa = 9.95RR262 pKa = 11.84VSNPMRR268 pKa = 11.84AEE270 pKa = 3.6QSVRR274 pKa = 11.84GFGVNPRR281 pKa = 11.84SVPRR285 pKa = 11.84VNPRR289 pKa = 11.84RR290 pKa = 11.84RR291 pKa = 11.84PPGVRR296 pKa = 11.84EE297 pKa = 3.95RR298 pKa = 11.84KK299 pKa = 9.35FKK301 pKa = 10.28LTISGSIVSNAFSFVTEE318 pKa = 4.1GADD321 pKa = 3.3VVEE324 pKa = 5.61AIYY327 pKa = 10.14EE328 pKa = 4.52AIPEE332 pKa = 4.52SIRR335 pKa = 11.84GTRR338 pKa = 11.84KK339 pKa = 10.1RR340 pKa = 11.84GVKK343 pKa = 8.7EE344 pKa = 3.15QAYY347 pKa = 8.6FIWQHH352 pKa = 5.98LSQVDD357 pKa = 3.8QNVMVQNLLYY367 pKa = 11.0NHH369 pKa = 6.71FEE371 pKa = 4.43DD372 pKa = 5.09KK373 pKa = 10.89FFGAIGEE380 pKa = 4.26RR381 pKa = 11.84MKK383 pKa = 10.74RR384 pKa = 11.84ANQVVWEE391 pKa = 4.04QGYY394 pKa = 11.02GNLRR398 pKa = 11.84VQFGLAHH405 pKa = 6.79
Molecular weight: 45.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1344 |
78 |
415 |
268.8 |
29.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.226 ± 0.576 | 1.19 ± 0.631 |
4.464 ± 1.155 | 5.283 ± 0.301 |
4.092 ± 0.592 | 7.738 ± 0.592 |
2.083 ± 0.309 | 3.274 ± 0.436 |
4.092 ± 0.784 | 7.813 ± 0.939 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.314 | 3.125 ± 0.507 |
8.185 ± 2.006 | 3.869 ± 0.494 |
8.78 ± 1.395 | 6.696 ± 0.738 |
4.911 ± 0.63 | 7.887 ± 0.555 |
2.009 ± 0.367 | 2.976 ± 0.32 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
