
Hubei sobemo-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
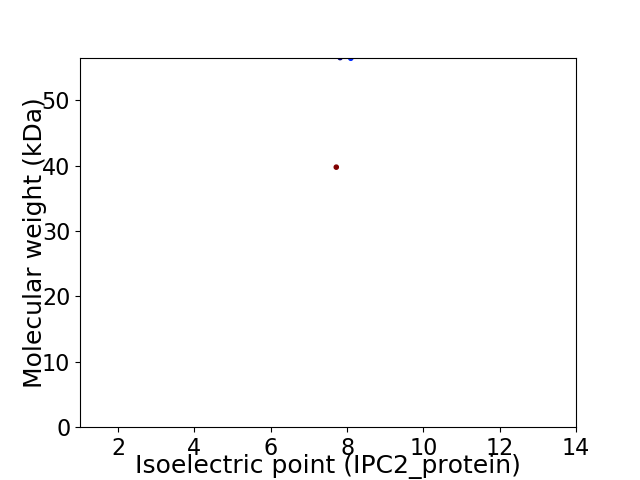
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KES8|A0A1L3KES8_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 9 OX=1923242 PE=4 SV=1
MM1 pKa = 8.23DD2 pKa = 4.41STPGNCQFKK11 pKa = 11.09SLGSTNKK18 pKa = 10.61DD19 pKa = 2.62IFKK22 pKa = 10.66YY23 pKa = 10.06EE24 pKa = 4.1AGICDD29 pKa = 4.17PEE31 pKa = 4.06RR32 pKa = 11.84VAMVRR37 pKa = 11.84CAVKK41 pKa = 10.6SRR43 pKa = 11.84LDD45 pKa = 3.55RR46 pKa = 11.84LIAGYY51 pKa = 10.69LEE53 pKa = 5.26ADD55 pKa = 4.15TINVFVKK62 pKa = 10.17PEE64 pKa = 3.73PHH66 pKa = 5.17TTAKK70 pKa = 9.33LTEE73 pKa = 4.03GRR75 pKa = 11.84LRR77 pKa = 11.84LISAVSLVDD86 pKa = 3.22TMIDD90 pKa = 2.76RR91 pKa = 11.84VLFGWVQRR99 pKa = 11.84KK100 pKa = 8.94ALTVVGRR107 pKa = 11.84TPCLCGWSPMVGGWRR122 pKa = 11.84YY123 pKa = 9.48IYY125 pKa = 10.84GRR127 pKa = 11.84FRR129 pKa = 11.84NQPVLCLDD137 pKa = 3.36KK138 pKa = 11.37SSWDD142 pKa = 3.27WTVQEE147 pKa = 4.72HH148 pKa = 6.56VVIFWKK154 pKa = 10.63RR155 pKa = 11.84FLSEE159 pKa = 3.76LAVGHH164 pKa = 6.0RR165 pKa = 11.84NWWTIAASARR175 pKa = 11.84FEE177 pKa = 4.38VLFKK181 pKa = 10.85LASFQFKK188 pKa = 10.59DD189 pKa = 3.46GTVVQQEE196 pKa = 4.32GTGIMKK202 pKa = 10.2SGCLLTLILNSVGQSYY218 pKa = 9.0LHH220 pKa = 5.94YY221 pKa = 10.82LSMLRR226 pKa = 11.84IGRR229 pKa = 11.84NPLQHH234 pKa = 6.51QPICVGDD241 pKa = 3.84DD242 pKa = 3.41TVQTSFEE249 pKa = 4.81DD250 pKa = 3.05IRR252 pKa = 11.84EE253 pKa = 4.08YY254 pKa = 10.65VYY256 pKa = 11.25EE257 pKa = 4.06MEE259 pKa = 4.58KK260 pKa = 10.75LGAVVKK266 pKa = 10.19GVKK269 pKa = 8.96IRR271 pKa = 11.84NWVEE275 pKa = 3.5FCGFAFAKK283 pKa = 6.95EE284 pKa = 4.45TCVPAYY290 pKa = 7.63WQKK293 pKa = 11.13HH294 pKa = 4.35LFKK297 pKa = 10.83LQYY300 pKa = 10.3CKK302 pKa = 11.01LEE304 pKa = 4.75DD305 pKa = 3.7SLASYY310 pKa = 10.42QMIYY314 pKa = 10.98ANEE317 pKa = 3.78PVMYY321 pKa = 10.44DD322 pKa = 3.4LLYY325 pKa = 10.76RR326 pKa = 11.84LAMQVNPEE334 pKa = 4.04LVITPVEE341 pKa = 4.23AKK343 pKa = 10.7LIMNGG348 pKa = 3.3
MM1 pKa = 8.23DD2 pKa = 4.41STPGNCQFKK11 pKa = 11.09SLGSTNKK18 pKa = 10.61DD19 pKa = 2.62IFKK22 pKa = 10.66YY23 pKa = 10.06EE24 pKa = 4.1AGICDD29 pKa = 4.17PEE31 pKa = 4.06RR32 pKa = 11.84VAMVRR37 pKa = 11.84CAVKK41 pKa = 10.6SRR43 pKa = 11.84LDD45 pKa = 3.55RR46 pKa = 11.84LIAGYY51 pKa = 10.69LEE53 pKa = 5.26ADD55 pKa = 4.15TINVFVKK62 pKa = 10.17PEE64 pKa = 3.73PHH66 pKa = 5.17TTAKK70 pKa = 9.33LTEE73 pKa = 4.03GRR75 pKa = 11.84LRR77 pKa = 11.84LISAVSLVDD86 pKa = 3.22TMIDD90 pKa = 2.76RR91 pKa = 11.84VLFGWVQRR99 pKa = 11.84KK100 pKa = 8.94ALTVVGRR107 pKa = 11.84TPCLCGWSPMVGGWRR122 pKa = 11.84YY123 pKa = 9.48IYY125 pKa = 10.84GRR127 pKa = 11.84FRR129 pKa = 11.84NQPVLCLDD137 pKa = 3.36KK138 pKa = 11.37SSWDD142 pKa = 3.27WTVQEE147 pKa = 4.72HH148 pKa = 6.56VVIFWKK154 pKa = 10.63RR155 pKa = 11.84FLSEE159 pKa = 3.76LAVGHH164 pKa = 6.0RR165 pKa = 11.84NWWTIAASARR175 pKa = 11.84FEE177 pKa = 4.38VLFKK181 pKa = 10.85LASFQFKK188 pKa = 10.59DD189 pKa = 3.46GTVVQQEE196 pKa = 4.32GTGIMKK202 pKa = 10.2SGCLLTLILNSVGQSYY218 pKa = 9.0LHH220 pKa = 5.94YY221 pKa = 10.82LSMLRR226 pKa = 11.84IGRR229 pKa = 11.84NPLQHH234 pKa = 6.51QPICVGDD241 pKa = 3.84DD242 pKa = 3.41TVQTSFEE249 pKa = 4.81DD250 pKa = 3.05IRR252 pKa = 11.84EE253 pKa = 4.08YY254 pKa = 10.65VYY256 pKa = 11.25EE257 pKa = 4.06MEE259 pKa = 4.58KK260 pKa = 10.75LGAVVKK266 pKa = 10.19GVKK269 pKa = 8.96IRR271 pKa = 11.84NWVEE275 pKa = 3.5FCGFAFAKK283 pKa = 6.95EE284 pKa = 4.45TCVPAYY290 pKa = 7.63WQKK293 pKa = 11.13HH294 pKa = 4.35LFKK297 pKa = 10.83LQYY300 pKa = 10.3CKK302 pKa = 11.01LEE304 pKa = 4.75DD305 pKa = 3.7SLASYY310 pKa = 10.42QMIYY314 pKa = 10.98ANEE317 pKa = 3.78PVMYY321 pKa = 10.44DD322 pKa = 3.4LLYY325 pKa = 10.76RR326 pKa = 11.84LAMQVNPEE334 pKa = 4.04LVITPVEE341 pKa = 4.23AKK343 pKa = 10.7LIMNGG348 pKa = 3.3
Molecular weight: 39.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KES8|A0A1L3KES8_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 9 OX=1923242 PE=4 SV=1
MM1 pKa = 7.06VFSEE5 pKa = 3.9ILMWSISSFYY15 pKa = 10.5AASFSSVMLVVIYY28 pKa = 8.89WHH30 pKa = 5.69LRR32 pKa = 11.84YY33 pKa = 8.71WRR35 pKa = 11.84KK36 pKa = 10.26RR37 pKa = 11.84IMRR40 pKa = 11.84NIMHH44 pKa = 6.84MWNLAEE50 pKa = 4.41YY51 pKa = 8.48YY52 pKa = 10.84VLMEE56 pKa = 4.23EE57 pKa = 4.43VEE59 pKa = 4.3PVYY62 pKa = 10.6QEE64 pKa = 4.07PKK66 pKa = 10.08GVMMDD71 pKa = 4.09FLEE74 pKa = 4.3TTYY77 pKa = 11.0GYY79 pKa = 10.59VVSKK83 pKa = 11.14LPGNSNLEE91 pKa = 3.81LFAYY95 pKa = 10.16VILSALGLIIAVFLLKK111 pKa = 10.72AIVKK115 pKa = 9.99SLMKK119 pKa = 10.29LYY121 pKa = 11.31VKK123 pKa = 9.6MQVYY127 pKa = 10.58DD128 pKa = 3.75LDD130 pKa = 4.11GFSEE134 pKa = 4.38KK135 pKa = 10.5VRR137 pKa = 11.84VGSVLEE143 pKa = 4.36NSMQEE148 pKa = 3.81PAFQAEE154 pKa = 4.07VWVRR158 pKa = 11.84RR159 pKa = 11.84KK160 pKa = 10.44GPYY163 pKa = 8.91VKK165 pKa = 10.19SGQGFLTNFGFFTAYY180 pKa = 10.11HH181 pKa = 4.99VVEE184 pKa = 4.57DD185 pKa = 4.31VEE187 pKa = 5.32SVMLVKK193 pKa = 10.65GSNKK197 pKa = 9.52VEE199 pKa = 3.78ITPDD203 pKa = 3.35LFVQLEE209 pKa = 4.27GDD211 pKa = 3.61VALYY215 pKa = 11.33KK216 pKa = 8.42MTQQVACKK224 pKa = 10.19LGLSQSKK231 pKa = 9.73LYY233 pKa = 10.14PIEE236 pKa = 4.48TPHH239 pKa = 7.24KK240 pKa = 8.12AGLVVRR246 pKa = 11.84VHH248 pKa = 6.99AFGQRR253 pKa = 11.84TMGVLRR259 pKa = 11.84PDD261 pKa = 3.17EE262 pKa = 4.62GFGLVVYY269 pKa = 9.84EE270 pKa = 4.35GSTIKK275 pKa = 10.68GFSGAPYY282 pKa = 8.8HH283 pKa = 6.48VGRR286 pKa = 11.84MVYY289 pKa = 9.74GMHH292 pKa = 6.47MGGSTVNLGLAGAYY306 pKa = 10.6LHH308 pKa = 6.48MLAKK312 pKa = 10.66YY313 pKa = 9.46EE314 pKa = 5.14LEE316 pKa = 4.33DD317 pKa = 3.61TDD319 pKa = 6.66DD320 pKa = 4.12YY321 pKa = 11.63LQQQIMNEE329 pKa = 3.84GKK331 pKa = 9.51EE332 pKa = 4.45FVWQRR337 pKa = 11.84SPYY340 pKa = 10.4DD341 pKa = 3.59PDD343 pKa = 3.25EE344 pKa = 4.23ARR346 pKa = 11.84VRR348 pKa = 11.84IEE350 pKa = 3.5GKK352 pKa = 10.54YY353 pKa = 9.5YY354 pKa = 8.56QTDD357 pKa = 2.94MDD359 pKa = 4.63FIRR362 pKa = 11.84RR363 pKa = 11.84MEE365 pKa = 3.97QKK367 pKa = 10.7VKK369 pKa = 10.53GKK371 pKa = 9.1QVSYY375 pKa = 10.71KK376 pKa = 10.27RR377 pKa = 11.84PDD379 pKa = 3.54YY380 pKa = 11.09EE381 pKa = 4.17EE382 pKa = 4.32EE383 pKa = 4.1ALSEE387 pKa = 4.27VSDD390 pKa = 3.98EE391 pKa = 4.58SLPLAPRR398 pKa = 11.84GAMSYY403 pKa = 10.47PDD405 pKa = 3.74SKK407 pKa = 11.66NLLEE411 pKa = 4.63APAVNAGASGKK422 pKa = 10.53LEE424 pKa = 3.84GRR426 pKa = 11.84VGVPKK431 pKa = 10.94GILNHH436 pKa = 6.0LRR438 pKa = 11.84EE439 pKa = 4.35IDD441 pKa = 3.76LEE443 pKa = 4.24SRR445 pKa = 11.84EE446 pKa = 4.24ALVSYY451 pKa = 10.79LMDD454 pKa = 4.19GQPVTRR460 pKa = 11.84VQQNEE465 pKa = 4.1VSGSTRR471 pKa = 11.84KK472 pKa = 9.84NSRR475 pKa = 11.84NRR477 pKa = 11.84KK478 pKa = 8.9RR479 pKa = 11.84KK480 pKa = 9.36LKK482 pKa = 10.52KK483 pKa = 9.32QVQRR487 pKa = 11.84QVKK490 pKa = 9.26QSSNNVV496 pKa = 3.04
MM1 pKa = 7.06VFSEE5 pKa = 3.9ILMWSISSFYY15 pKa = 10.5AASFSSVMLVVIYY28 pKa = 8.89WHH30 pKa = 5.69LRR32 pKa = 11.84YY33 pKa = 8.71WRR35 pKa = 11.84KK36 pKa = 10.26RR37 pKa = 11.84IMRR40 pKa = 11.84NIMHH44 pKa = 6.84MWNLAEE50 pKa = 4.41YY51 pKa = 8.48YY52 pKa = 10.84VLMEE56 pKa = 4.23EE57 pKa = 4.43VEE59 pKa = 4.3PVYY62 pKa = 10.6QEE64 pKa = 4.07PKK66 pKa = 10.08GVMMDD71 pKa = 4.09FLEE74 pKa = 4.3TTYY77 pKa = 11.0GYY79 pKa = 10.59VVSKK83 pKa = 11.14LPGNSNLEE91 pKa = 3.81LFAYY95 pKa = 10.16VILSALGLIIAVFLLKK111 pKa = 10.72AIVKK115 pKa = 9.99SLMKK119 pKa = 10.29LYY121 pKa = 11.31VKK123 pKa = 9.6MQVYY127 pKa = 10.58DD128 pKa = 3.75LDD130 pKa = 4.11GFSEE134 pKa = 4.38KK135 pKa = 10.5VRR137 pKa = 11.84VGSVLEE143 pKa = 4.36NSMQEE148 pKa = 3.81PAFQAEE154 pKa = 4.07VWVRR158 pKa = 11.84RR159 pKa = 11.84KK160 pKa = 10.44GPYY163 pKa = 8.91VKK165 pKa = 10.19SGQGFLTNFGFFTAYY180 pKa = 10.11HH181 pKa = 4.99VVEE184 pKa = 4.57DD185 pKa = 4.31VEE187 pKa = 5.32SVMLVKK193 pKa = 10.65GSNKK197 pKa = 9.52VEE199 pKa = 3.78ITPDD203 pKa = 3.35LFVQLEE209 pKa = 4.27GDD211 pKa = 3.61VALYY215 pKa = 11.33KK216 pKa = 8.42MTQQVACKK224 pKa = 10.19LGLSQSKK231 pKa = 9.73LYY233 pKa = 10.14PIEE236 pKa = 4.48TPHH239 pKa = 7.24KK240 pKa = 8.12AGLVVRR246 pKa = 11.84VHH248 pKa = 6.99AFGQRR253 pKa = 11.84TMGVLRR259 pKa = 11.84PDD261 pKa = 3.17EE262 pKa = 4.62GFGLVVYY269 pKa = 9.84EE270 pKa = 4.35GSTIKK275 pKa = 10.68GFSGAPYY282 pKa = 8.8HH283 pKa = 6.48VGRR286 pKa = 11.84MVYY289 pKa = 9.74GMHH292 pKa = 6.47MGGSTVNLGLAGAYY306 pKa = 10.6LHH308 pKa = 6.48MLAKK312 pKa = 10.66YY313 pKa = 9.46EE314 pKa = 5.14LEE316 pKa = 4.33DD317 pKa = 3.61TDD319 pKa = 6.66DD320 pKa = 4.12YY321 pKa = 11.63LQQQIMNEE329 pKa = 3.84GKK331 pKa = 9.51EE332 pKa = 4.45FVWQRR337 pKa = 11.84SPYY340 pKa = 10.4DD341 pKa = 3.59PDD343 pKa = 3.25EE344 pKa = 4.23ARR346 pKa = 11.84VRR348 pKa = 11.84IEE350 pKa = 3.5GKK352 pKa = 10.54YY353 pKa = 9.5YY354 pKa = 8.56QTDD357 pKa = 2.94MDD359 pKa = 4.63FIRR362 pKa = 11.84RR363 pKa = 11.84MEE365 pKa = 3.97QKK367 pKa = 10.7VKK369 pKa = 10.53GKK371 pKa = 9.1QVSYY375 pKa = 10.71KK376 pKa = 10.27RR377 pKa = 11.84PDD379 pKa = 3.54YY380 pKa = 11.09EE381 pKa = 4.17EE382 pKa = 4.32EE383 pKa = 4.1ALSEE387 pKa = 4.27VSDD390 pKa = 3.98EE391 pKa = 4.58SLPLAPRR398 pKa = 11.84GAMSYY403 pKa = 10.47PDD405 pKa = 3.74SKK407 pKa = 11.66NLLEE411 pKa = 4.63APAVNAGASGKK422 pKa = 10.53LEE424 pKa = 3.84GRR426 pKa = 11.84VGVPKK431 pKa = 10.94GILNHH436 pKa = 6.0LRR438 pKa = 11.84EE439 pKa = 4.35IDD441 pKa = 3.76LEE443 pKa = 4.24SRR445 pKa = 11.84EE446 pKa = 4.24ALVSYY451 pKa = 10.79LMDD454 pKa = 4.19GQPVTRR460 pKa = 11.84VQQNEE465 pKa = 4.1VSGSTRR471 pKa = 11.84KK472 pKa = 9.84NSRR475 pKa = 11.84NRR477 pKa = 11.84KK478 pKa = 8.9RR479 pKa = 11.84KK480 pKa = 9.36LKK482 pKa = 10.52KK483 pKa = 9.32QVQRR487 pKa = 11.84QVKK490 pKa = 9.26QSSNNVV496 pKa = 3.04
Molecular weight: 56.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
844 |
348 |
496 |
422.0 |
48.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.687 ± 0.216 | 1.422 ± 1.081 |
4.028 ± 0.175 | 6.635 ± 0.731 |
4.028 ± 0.354 | 7.227 ± 0.384 |
1.777 ± 0.033 | 4.147 ± 0.638 |
6.28 ± 0.331 | 9.716 ± 0.213 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.147 ± 0.613 | 3.318 ± 0.097 |
3.791 ± 0.035 | 4.621 ± 0.193 |
5.569 ± 0.111 | 6.635 ± 0.731 |
3.791 ± 0.859 | 10.308 ± 0.513 |
1.896 ± 0.608 | 4.976 ± 0.593 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
