
Microvirga brassicacearum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Methylobacteriaceae; Microvirga
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
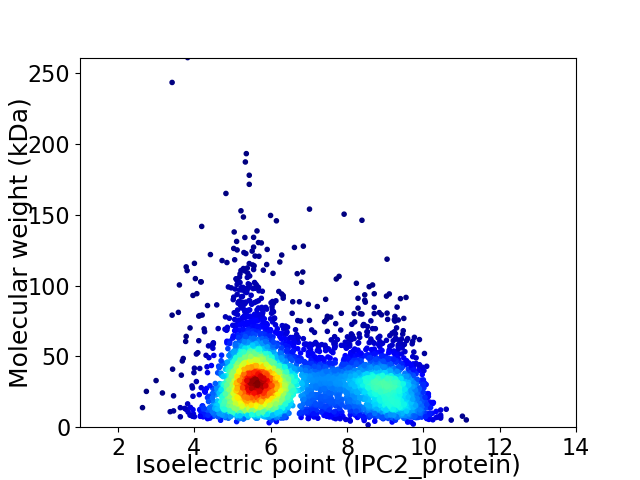
Virtual 2D-PAGE plot for 4754 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N3PDC2|A0A5N3PDC2_9RHIZ Response regulator transcription factor OS=Microvirga brassicacearum OX=2580413 GN=FEZ63_10705 PE=4 SV=1
MM1 pKa = 7.84AIGSSIEE8 pKa = 4.48LIWHH12 pKa = 5.85FAGYY16 pKa = 9.98LHH18 pKa = 7.12LTQDD22 pKa = 3.93YY23 pKa = 8.39VASRR27 pKa = 11.84VRR29 pKa = 11.84LDD31 pKa = 4.5EE32 pKa = 5.28IIHH35 pKa = 6.28SNDD38 pKa = 2.65TAADD42 pKa = 3.77FRR44 pKa = 11.84VSADD48 pKa = 3.22EE49 pKa = 3.75YY50 pKa = 10.21LARR53 pKa = 11.84SEE55 pKa = 4.94APSDD59 pKa = 3.38MSNFAIQVKK68 pKa = 10.16YY69 pKa = 11.05SLGSEE74 pKa = 3.98NPDD77 pKa = 3.11RR78 pKa = 11.84GEE80 pKa = 3.96WADD83 pKa = 5.18LPDD86 pKa = 5.9DD87 pKa = 5.08DD88 pKa = 5.43SLDD91 pKa = 3.59VGAPNVDD98 pKa = 4.83LLPHH102 pKa = 6.77DD103 pKa = 4.16WSTGTSSLFAGQSYY117 pKa = 9.24VVSCTAEE124 pKa = 3.76SSGPGGSHH132 pKa = 6.37LQDD135 pKa = 3.96SQSHH139 pKa = 6.04PASTYY144 pKa = 8.42EE145 pKa = 4.31SKK147 pKa = 10.98SADD150 pKa = 3.55GGDD153 pKa = 3.44DD154 pKa = 3.8TIANLSQANIGADD167 pKa = 2.88RR168 pKa = 11.84DD169 pKa = 4.16YY170 pKa = 11.34FSDD173 pKa = 3.93GPNEE177 pKa = 4.42LGVLPHH183 pKa = 6.17VASDD187 pKa = 3.66QPAADD192 pKa = 3.76MLTLAEE198 pKa = 4.73EE199 pKa = 4.49YY200 pKa = 10.75NPTTMLPPGEE210 pKa = 4.64SVEE213 pKa = 4.4FWTSAVVDD221 pKa = 3.73RR222 pKa = 11.84HH223 pKa = 5.58EE224 pKa = 4.39SLAAGEE230 pKa = 4.73TEE232 pKa = 4.29AQTIAPGRR240 pKa = 11.84YY241 pKa = 8.78VNGKK245 pKa = 9.07LVALEE250 pKa = 4.36PSVEE254 pKa = 4.15TAQPANHH261 pKa = 6.97TLPEE265 pKa = 4.25PPVRR269 pKa = 11.84EE270 pKa = 4.2VGSTDD275 pKa = 3.25PAQIAEE281 pKa = 4.27TGGNKK286 pKa = 9.38LVNAATIADD295 pKa = 4.3LNEE298 pKa = 4.28APSTLIVGGDD308 pKa = 3.69YY309 pKa = 11.47YY310 pKa = 10.25EE311 pKa = 4.6TNAILQTNILQNQDD325 pKa = 3.38VVFHH329 pKa = 7.37DD330 pKa = 4.21GDD332 pKa = 4.33APPMLDD338 pKa = 3.27STGSTLDD345 pKa = 3.23NVANFVVEE353 pKa = 4.35EE354 pKa = 4.03LVKK357 pKa = 10.76QQGHH361 pKa = 5.84KK362 pKa = 10.59AGPLGNLTVNIDD374 pKa = 3.77YY375 pKa = 11.28VDD377 pKa = 3.62GHH379 pKa = 7.49VMDD382 pKa = 5.14VKK384 pKa = 11.17ALAQRR389 pKa = 11.84NYY391 pKa = 10.54FEE393 pKa = 6.16DD394 pKa = 4.51GDD396 pKa = 4.15VTVQTRR402 pKa = 11.84YY403 pKa = 10.55DD404 pKa = 3.85SYY406 pKa = 11.93SEE408 pKa = 3.74IHH410 pKa = 6.8SGDD413 pKa = 3.19NTQANVARR421 pKa = 11.84FADD424 pKa = 3.26WGKK427 pKa = 10.55DD428 pKa = 3.26YY429 pKa = 11.51DD430 pKa = 4.39VIIVMGDD437 pKa = 3.2YY438 pKa = 10.83HH439 pKa = 6.84SANLILQTNVVLDD452 pKa = 4.08DD453 pKa = 4.58DD454 pKa = 5.26YY455 pKa = 11.83IGISCGSGGSTPSIFSGQNALEE477 pKa = 4.21NEE479 pKa = 4.07ATIAKK484 pKa = 10.02YY485 pKa = 10.38GATTFGGINRR495 pKa = 11.84KK496 pKa = 9.24LDD498 pKa = 3.7SLIDD502 pKa = 4.08DD503 pKa = 4.74LNDD506 pKa = 4.48RR507 pKa = 11.84DD508 pKa = 4.94DD509 pKa = 4.76LDD511 pKa = 4.26RR512 pKa = 11.84NAWSDD517 pKa = 3.46FHH519 pKa = 6.54GASSGSLNVLFVTGNYY535 pKa = 9.72YY536 pKa = 10.67DD537 pKa = 4.98LNIISQINVLADD549 pKa = 3.36ADD551 pKa = 4.12FAIQGGSDD559 pKa = 2.73GWQWLSTGGNSALNQASIVNAGGVYY584 pKa = 9.65DD585 pKa = 3.54QHH587 pKa = 8.46LGGDD591 pKa = 4.34FYY593 pKa = 11.45EE594 pKa = 5.52DD595 pKa = 3.87SILVQANLMSDD606 pKa = 3.33GSEE609 pKa = 4.05AVATDD614 pKa = 3.55PTALVSEE621 pKa = 4.89LVAFMDD627 pKa = 3.68HH628 pKa = 6.3TPDD631 pKa = 3.59YY632 pKa = 10.6SDD634 pKa = 3.91SDD636 pKa = 4.15SVSWTKK642 pKa = 10.39DD643 pKa = 3.11TFGHH647 pKa = 6.92GDD649 pKa = 3.36TFGHH653 pKa = 5.89VLSS656 pKa = 4.75
MM1 pKa = 7.84AIGSSIEE8 pKa = 4.48LIWHH12 pKa = 5.85FAGYY16 pKa = 9.98LHH18 pKa = 7.12LTQDD22 pKa = 3.93YY23 pKa = 8.39VASRR27 pKa = 11.84VRR29 pKa = 11.84LDD31 pKa = 4.5EE32 pKa = 5.28IIHH35 pKa = 6.28SNDD38 pKa = 2.65TAADD42 pKa = 3.77FRR44 pKa = 11.84VSADD48 pKa = 3.22EE49 pKa = 3.75YY50 pKa = 10.21LARR53 pKa = 11.84SEE55 pKa = 4.94APSDD59 pKa = 3.38MSNFAIQVKK68 pKa = 10.16YY69 pKa = 11.05SLGSEE74 pKa = 3.98NPDD77 pKa = 3.11RR78 pKa = 11.84GEE80 pKa = 3.96WADD83 pKa = 5.18LPDD86 pKa = 5.9DD87 pKa = 5.08DD88 pKa = 5.43SLDD91 pKa = 3.59VGAPNVDD98 pKa = 4.83LLPHH102 pKa = 6.77DD103 pKa = 4.16WSTGTSSLFAGQSYY117 pKa = 9.24VVSCTAEE124 pKa = 3.76SSGPGGSHH132 pKa = 6.37LQDD135 pKa = 3.96SQSHH139 pKa = 6.04PASTYY144 pKa = 8.42EE145 pKa = 4.31SKK147 pKa = 10.98SADD150 pKa = 3.55GGDD153 pKa = 3.44DD154 pKa = 3.8TIANLSQANIGADD167 pKa = 2.88RR168 pKa = 11.84DD169 pKa = 4.16YY170 pKa = 11.34FSDD173 pKa = 3.93GPNEE177 pKa = 4.42LGVLPHH183 pKa = 6.17VASDD187 pKa = 3.66QPAADD192 pKa = 3.76MLTLAEE198 pKa = 4.73EE199 pKa = 4.49YY200 pKa = 10.75NPTTMLPPGEE210 pKa = 4.64SVEE213 pKa = 4.4FWTSAVVDD221 pKa = 3.73RR222 pKa = 11.84HH223 pKa = 5.58EE224 pKa = 4.39SLAAGEE230 pKa = 4.73TEE232 pKa = 4.29AQTIAPGRR240 pKa = 11.84YY241 pKa = 8.78VNGKK245 pKa = 9.07LVALEE250 pKa = 4.36PSVEE254 pKa = 4.15TAQPANHH261 pKa = 6.97TLPEE265 pKa = 4.25PPVRR269 pKa = 11.84EE270 pKa = 4.2VGSTDD275 pKa = 3.25PAQIAEE281 pKa = 4.27TGGNKK286 pKa = 9.38LVNAATIADD295 pKa = 4.3LNEE298 pKa = 4.28APSTLIVGGDD308 pKa = 3.69YY309 pKa = 11.47YY310 pKa = 10.25EE311 pKa = 4.6TNAILQTNILQNQDD325 pKa = 3.38VVFHH329 pKa = 7.37DD330 pKa = 4.21GDD332 pKa = 4.33APPMLDD338 pKa = 3.27STGSTLDD345 pKa = 3.23NVANFVVEE353 pKa = 4.35EE354 pKa = 4.03LVKK357 pKa = 10.76QQGHH361 pKa = 5.84KK362 pKa = 10.59AGPLGNLTVNIDD374 pKa = 3.77YY375 pKa = 11.28VDD377 pKa = 3.62GHH379 pKa = 7.49VMDD382 pKa = 5.14VKK384 pKa = 11.17ALAQRR389 pKa = 11.84NYY391 pKa = 10.54FEE393 pKa = 6.16DD394 pKa = 4.51GDD396 pKa = 4.15VTVQTRR402 pKa = 11.84YY403 pKa = 10.55DD404 pKa = 3.85SYY406 pKa = 11.93SEE408 pKa = 3.74IHH410 pKa = 6.8SGDD413 pKa = 3.19NTQANVARR421 pKa = 11.84FADD424 pKa = 3.26WGKK427 pKa = 10.55DD428 pKa = 3.26YY429 pKa = 11.51DD430 pKa = 4.39VIIVMGDD437 pKa = 3.2YY438 pKa = 10.83HH439 pKa = 6.84SANLILQTNVVLDD452 pKa = 4.08DD453 pKa = 4.58DD454 pKa = 5.26YY455 pKa = 11.83IGISCGSGGSTPSIFSGQNALEE477 pKa = 4.21NEE479 pKa = 4.07ATIAKK484 pKa = 10.02YY485 pKa = 10.38GATTFGGINRR495 pKa = 11.84KK496 pKa = 9.24LDD498 pKa = 3.7SLIDD502 pKa = 4.08DD503 pKa = 4.74LNDD506 pKa = 4.48RR507 pKa = 11.84DD508 pKa = 4.94DD509 pKa = 4.76LDD511 pKa = 4.26RR512 pKa = 11.84NAWSDD517 pKa = 3.46FHH519 pKa = 6.54GASSGSLNVLFVTGNYY535 pKa = 9.72YY536 pKa = 10.67DD537 pKa = 4.98LNIISQINVLADD549 pKa = 3.36ADD551 pKa = 4.12FAIQGGSDD559 pKa = 2.73GWQWLSTGGNSALNQASIVNAGGVYY584 pKa = 9.65DD585 pKa = 3.54QHH587 pKa = 8.46LGGDD591 pKa = 4.34FYY593 pKa = 11.45EE594 pKa = 5.52DD595 pKa = 3.87SILVQANLMSDD606 pKa = 3.33GSEE609 pKa = 4.05AVATDD614 pKa = 3.55PTALVSEE621 pKa = 4.89LVAFMDD627 pKa = 3.68HH628 pKa = 6.3TPDD631 pKa = 3.59YY632 pKa = 10.6SDD634 pKa = 3.91SDD636 pKa = 4.15SVSWTKK642 pKa = 10.39DD643 pKa = 3.11TFGHH647 pKa = 6.92GDD649 pKa = 3.36TFGHH653 pKa = 5.89VLSS656 pKa = 4.75
Molecular weight: 70.1 kDa
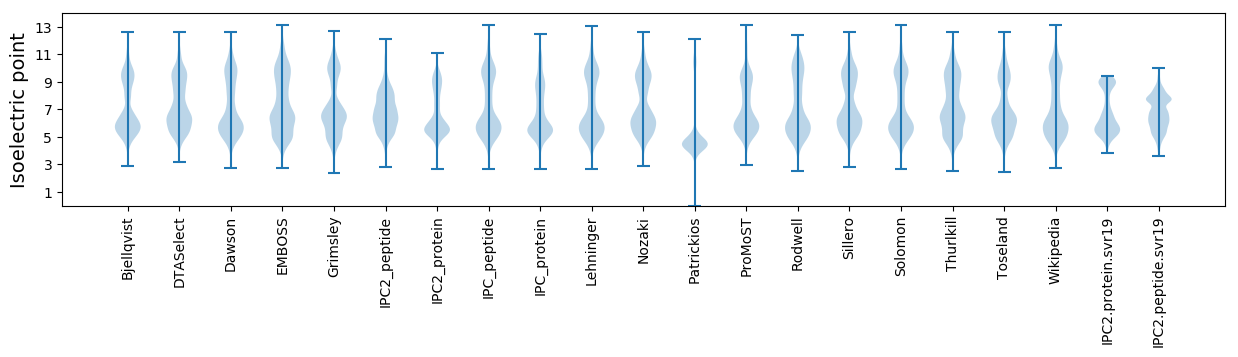
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N3PD11|A0A5N3PD11_9RHIZ 2 3 4 5-tetrahydropyridine-2 6-dicarboxylate N-succinyltransferase OS=Microvirga brassicacearum OX=2580413 GN=dapD PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.11GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.38GGRR28 pKa = 11.84KK29 pKa = 9.21VIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.11GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497474 |
16 |
2501 |
315.0 |
34.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.043 ± 0.048 | 0.785 ± 0.011 |
5.645 ± 0.029 | 5.547 ± 0.029 |
3.815 ± 0.021 | 8.595 ± 0.038 |
1.995 ± 0.017 | 5.443 ± 0.023 |
3.239 ± 0.033 | 10.237 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.38 ± 0.016 | 2.655 ± 0.024 |
5.215 ± 0.028 | 3.027 ± 0.019 |
7.253 ± 0.036 | 5.618 ± 0.022 |
5.405 ± 0.027 | 7.593 ± 0.028 |
1.309 ± 0.015 | 2.199 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
