
Kordiimonas lacus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Kordiimonadales; Kordiimonadaceae; Kordiimonas
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
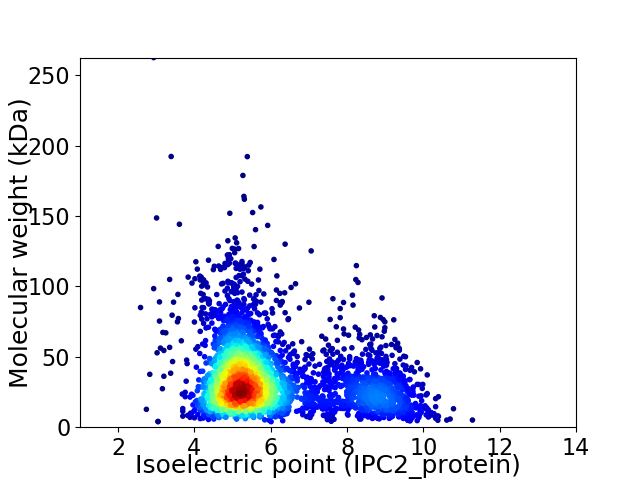
Virtual 2D-PAGE plot for 3653 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6XKV3|A0A1G6XKV3_9PROT NAD(+) diphosphatase OS=Kordiimonas lacus OX=637679 GN=SAMN04488071_1277 PE=4 SV=1
MM1 pKa = 7.64GNTGTDD7 pKa = 2.9KK8 pKa = 10.95AAPQTADD15 pKa = 3.64DD16 pKa = 4.29TGDD19 pKa = 3.55GYY21 pKa = 9.29TIPFGDD27 pKa = 3.56FAGTYY32 pKa = 10.35VPFNDD37 pKa = 4.26PLFPFQANLTGFMYY51 pKa = 10.6DD52 pKa = 3.12DD53 pKa = 3.52TGAYY57 pKa = 8.52TEE59 pKa = 4.45SAHH62 pKa = 7.61INILPVWDD70 pKa = 4.84EE71 pKa = 3.96YY72 pKa = 11.63DD73 pKa = 3.35GTGILIASVEE83 pKa = 4.53EE84 pKa = 3.74IDD86 pKa = 5.14FRR88 pKa = 11.84HH89 pKa = 6.69PDD91 pKa = 3.33LEE93 pKa = 4.46GAQVDD98 pKa = 4.1NAPVSYY104 pKa = 11.35EE105 pKa = 3.85EE106 pKa = 4.04MFQNIQHH113 pKa = 7.37DD114 pKa = 4.61YY115 pKa = 9.04THH117 pKa = 6.39GTSTAGIIGARR128 pKa = 11.84AGNGIGLAGVAHH140 pKa = 7.13GSSILAGYY148 pKa = 9.56SIYY151 pKa = 11.18NGVTAEE157 pKa = 4.37LNADD161 pKa = 3.18VDD163 pKa = 3.76IVTRR167 pKa = 11.84SVSGGFLLSSDD178 pKa = 4.19LNGGYY183 pKa = 10.04SGATDD188 pKa = 4.12FLAEE192 pKa = 4.22GRR194 pKa = 11.84DD195 pKa = 3.6GLGIVWVNGSGNWRR209 pKa = 11.84YY210 pKa = 10.34NDD212 pKa = 3.37STTDD216 pKa = 3.26ISWGLNEE223 pKa = 5.52LYY225 pKa = 10.65VLSVGAASPNGFVTVFSVAGSALHH249 pKa = 5.83MVSPLAQAFLGAKK262 pKa = 9.85SDD264 pKa = 3.82TTLSLDD270 pKa = 3.94RR271 pKa = 11.84PGNWGEE277 pKa = 3.87IKK279 pKa = 10.51ILGAEE284 pKa = 3.78ITEE287 pKa = 4.28FEE289 pKa = 4.34ITGQAPQEE297 pKa = 4.0PGFYY301 pKa = 10.2LQVVEE306 pKa = 5.06NSLAHH311 pKa = 6.52AALEE315 pKa = 4.3AGFEE319 pKa = 4.31DD320 pKa = 4.49GDD322 pKa = 4.2YY323 pKa = 10.77IFYY326 pKa = 10.9GGTSSSGPLVSGATALVLEE345 pKa = 4.62AASNNVFGDD354 pKa = 3.91TEE356 pKa = 4.62LGWRR360 pKa = 11.84DD361 pKa = 3.44VQEE364 pKa = 4.08IFALSSAHH372 pKa = 6.04TGSGFGITEE381 pKa = 3.91RR382 pKa = 11.84DD383 pKa = 3.79LYY385 pKa = 11.28DD386 pKa = 4.15HH387 pKa = 7.14EE388 pKa = 4.86LNSWIINGASILNGGGLHH406 pKa = 6.63WSADD410 pKa = 3.7YY411 pKa = 11.12GFGMLDD417 pKa = 2.49VHH419 pKa = 6.46AAVRR423 pKa = 11.84LAEE426 pKa = 4.12TWTMTRR432 pKa = 11.84HH433 pKa = 5.58SGNLVEE439 pKa = 4.68QSHH442 pKa = 6.01SFATASEE449 pKa = 4.49TITYY453 pKa = 9.93GADD456 pKa = 3.02LTFKK460 pKa = 10.51FVAGDD465 pKa = 3.61ADD467 pKa = 4.87AIDD470 pKa = 3.56IDD472 pKa = 4.47GVEE475 pKa = 3.96LQFDD479 pKa = 4.06IVHH482 pKa = 6.68DD483 pKa = 4.22AWPEE487 pKa = 3.74LEE489 pKa = 5.0MILISPSGTEE499 pKa = 3.95IILFDD504 pKa = 4.15TPEE507 pKa = 4.0LTASAPHH514 pKa = 6.51EE515 pKa = 4.33SANTGQLEE523 pKa = 4.1WDD525 pKa = 3.66LLARR529 pKa = 11.84GFWGEE534 pKa = 3.86EE535 pKa = 4.07SAGEE539 pKa = 3.93WTLVIRR545 pKa = 11.84DD546 pKa = 3.83TVDD549 pKa = 2.78NGNSGTVNSLSLMFLGDD566 pKa = 3.55AATDD570 pKa = 3.45DD571 pKa = 4.77DD572 pKa = 5.0IYY574 pKa = 11.53YY575 pKa = 8.22FTDD578 pKa = 3.54DD579 pKa = 3.56WALMNAANGGVPVLDD594 pKa = 4.84DD595 pKa = 4.59ALGQNSINAAALMEE609 pKa = 5.31AILLKK614 pKa = 10.36LAPGSTSKK622 pKa = 10.55IGGEE626 pKa = 4.21TAFKK630 pKa = 10.12TSADD634 pKa = 3.38AVFTLGIGTDD644 pKa = 3.4QDD646 pKa = 3.73DD647 pKa = 4.57TLVGAAFDD655 pKa = 3.61EE656 pKa = 4.68TLYY659 pKa = 11.39GMQGDD664 pKa = 3.86DD665 pKa = 4.85RR666 pKa = 11.84MWAGAGDD673 pKa = 3.88VGADD677 pKa = 3.41SYY679 pKa = 11.89YY680 pKa = 11.11GGDD683 pKa = 3.53GADD686 pKa = 3.52IIGVGAGNDD695 pKa = 3.77LAVGEE700 pKa = 4.94AGNDD704 pKa = 3.91AIFGGAGDD712 pKa = 4.01DD713 pKa = 3.84TLVGGFWEE721 pKa = 5.32GGSASAVGGGHH732 pKa = 6.06NEE734 pKa = 3.63LWAGSGDD741 pKa = 4.39DD742 pKa = 4.6VIHH745 pKa = 6.55GANGNDD751 pKa = 3.33ALGGGTEE758 pKa = 4.09SDD760 pKa = 3.52TIYY763 pKa = 10.64GYY765 pKa = 10.79GGHH768 pKa = 7.76DD769 pKa = 2.99ILYY772 pKa = 8.99AGKK775 pKa = 10.25GAVSDD780 pKa = 4.2TLDD783 pKa = 3.64GGAGQDD789 pKa = 3.45TLFGGGGDD797 pKa = 4.26DD798 pKa = 3.67MLSGGGGNDD807 pKa = 4.71LIFNGAGSDD816 pKa = 3.53TATGGDD822 pKa = 4.1GDD824 pKa = 3.95DD825 pKa = 4.15TLWGGGGDD833 pKa = 4.14DD834 pKa = 5.25LLTGGTGADD843 pKa = 2.65IFAFTSTNGSDD854 pKa = 2.98TVTDD858 pKa = 4.08FDD860 pKa = 3.91AAEE863 pKa = 5.51DD864 pKa = 3.59ILLLDD869 pKa = 4.17GSGFADD875 pKa = 4.74LAALKK880 pKa = 10.44AATTEE885 pKa = 4.25DD886 pKa = 3.82GAGVTISFGDD896 pKa = 3.55TVIILSGISLDD907 pKa = 4.71DD908 pKa = 3.38LTAANVMLL916 pKa = 4.98
MM1 pKa = 7.64GNTGTDD7 pKa = 2.9KK8 pKa = 10.95AAPQTADD15 pKa = 3.64DD16 pKa = 4.29TGDD19 pKa = 3.55GYY21 pKa = 9.29TIPFGDD27 pKa = 3.56FAGTYY32 pKa = 10.35VPFNDD37 pKa = 4.26PLFPFQANLTGFMYY51 pKa = 10.6DD52 pKa = 3.12DD53 pKa = 3.52TGAYY57 pKa = 8.52TEE59 pKa = 4.45SAHH62 pKa = 7.61INILPVWDD70 pKa = 4.84EE71 pKa = 3.96YY72 pKa = 11.63DD73 pKa = 3.35GTGILIASVEE83 pKa = 4.53EE84 pKa = 3.74IDD86 pKa = 5.14FRR88 pKa = 11.84HH89 pKa = 6.69PDD91 pKa = 3.33LEE93 pKa = 4.46GAQVDD98 pKa = 4.1NAPVSYY104 pKa = 11.35EE105 pKa = 3.85EE106 pKa = 4.04MFQNIQHH113 pKa = 7.37DD114 pKa = 4.61YY115 pKa = 9.04THH117 pKa = 6.39GTSTAGIIGARR128 pKa = 11.84AGNGIGLAGVAHH140 pKa = 7.13GSSILAGYY148 pKa = 9.56SIYY151 pKa = 11.18NGVTAEE157 pKa = 4.37LNADD161 pKa = 3.18VDD163 pKa = 3.76IVTRR167 pKa = 11.84SVSGGFLLSSDD178 pKa = 4.19LNGGYY183 pKa = 10.04SGATDD188 pKa = 4.12FLAEE192 pKa = 4.22GRR194 pKa = 11.84DD195 pKa = 3.6GLGIVWVNGSGNWRR209 pKa = 11.84YY210 pKa = 10.34NDD212 pKa = 3.37STTDD216 pKa = 3.26ISWGLNEE223 pKa = 5.52LYY225 pKa = 10.65VLSVGAASPNGFVTVFSVAGSALHH249 pKa = 5.83MVSPLAQAFLGAKK262 pKa = 9.85SDD264 pKa = 3.82TTLSLDD270 pKa = 3.94RR271 pKa = 11.84PGNWGEE277 pKa = 3.87IKK279 pKa = 10.51ILGAEE284 pKa = 3.78ITEE287 pKa = 4.28FEE289 pKa = 4.34ITGQAPQEE297 pKa = 4.0PGFYY301 pKa = 10.2LQVVEE306 pKa = 5.06NSLAHH311 pKa = 6.52AALEE315 pKa = 4.3AGFEE319 pKa = 4.31DD320 pKa = 4.49GDD322 pKa = 4.2YY323 pKa = 10.77IFYY326 pKa = 10.9GGTSSSGPLVSGATALVLEE345 pKa = 4.62AASNNVFGDD354 pKa = 3.91TEE356 pKa = 4.62LGWRR360 pKa = 11.84DD361 pKa = 3.44VQEE364 pKa = 4.08IFALSSAHH372 pKa = 6.04TGSGFGITEE381 pKa = 3.91RR382 pKa = 11.84DD383 pKa = 3.79LYY385 pKa = 11.28DD386 pKa = 4.15HH387 pKa = 7.14EE388 pKa = 4.86LNSWIINGASILNGGGLHH406 pKa = 6.63WSADD410 pKa = 3.7YY411 pKa = 11.12GFGMLDD417 pKa = 2.49VHH419 pKa = 6.46AAVRR423 pKa = 11.84LAEE426 pKa = 4.12TWTMTRR432 pKa = 11.84HH433 pKa = 5.58SGNLVEE439 pKa = 4.68QSHH442 pKa = 6.01SFATASEE449 pKa = 4.49TITYY453 pKa = 9.93GADD456 pKa = 3.02LTFKK460 pKa = 10.51FVAGDD465 pKa = 3.61ADD467 pKa = 4.87AIDD470 pKa = 3.56IDD472 pKa = 4.47GVEE475 pKa = 3.96LQFDD479 pKa = 4.06IVHH482 pKa = 6.68DD483 pKa = 4.22AWPEE487 pKa = 3.74LEE489 pKa = 5.0MILISPSGTEE499 pKa = 3.95IILFDD504 pKa = 4.15TPEE507 pKa = 4.0LTASAPHH514 pKa = 6.51EE515 pKa = 4.33SANTGQLEE523 pKa = 4.1WDD525 pKa = 3.66LLARR529 pKa = 11.84GFWGEE534 pKa = 3.86EE535 pKa = 4.07SAGEE539 pKa = 3.93WTLVIRR545 pKa = 11.84DD546 pKa = 3.83TVDD549 pKa = 2.78NGNSGTVNSLSLMFLGDD566 pKa = 3.55AATDD570 pKa = 3.45DD571 pKa = 4.77DD572 pKa = 5.0IYY574 pKa = 11.53YY575 pKa = 8.22FTDD578 pKa = 3.54DD579 pKa = 3.56WALMNAANGGVPVLDD594 pKa = 4.84DD595 pKa = 4.59ALGQNSINAAALMEE609 pKa = 5.31AILLKK614 pKa = 10.36LAPGSTSKK622 pKa = 10.55IGGEE626 pKa = 4.21TAFKK630 pKa = 10.12TSADD634 pKa = 3.38AVFTLGIGTDD644 pKa = 3.4QDD646 pKa = 3.73DD647 pKa = 4.57TLVGAAFDD655 pKa = 3.61EE656 pKa = 4.68TLYY659 pKa = 11.39GMQGDD664 pKa = 3.86DD665 pKa = 4.85RR666 pKa = 11.84MWAGAGDD673 pKa = 3.88VGADD677 pKa = 3.41SYY679 pKa = 11.89YY680 pKa = 11.11GGDD683 pKa = 3.53GADD686 pKa = 3.52IIGVGAGNDD695 pKa = 3.77LAVGEE700 pKa = 4.94AGNDD704 pKa = 3.91AIFGGAGDD712 pKa = 4.01DD713 pKa = 3.84TLVGGFWEE721 pKa = 5.32GGSASAVGGGHH732 pKa = 6.06NEE734 pKa = 3.63LWAGSGDD741 pKa = 4.39DD742 pKa = 4.6VIHH745 pKa = 6.55GANGNDD751 pKa = 3.33ALGGGTEE758 pKa = 4.09SDD760 pKa = 3.52TIYY763 pKa = 10.64GYY765 pKa = 10.79GGHH768 pKa = 7.76DD769 pKa = 2.99ILYY772 pKa = 8.99AGKK775 pKa = 10.25GAVSDD780 pKa = 4.2TLDD783 pKa = 3.64GGAGQDD789 pKa = 3.45TLFGGGGDD797 pKa = 4.26DD798 pKa = 3.67MLSGGGGNDD807 pKa = 4.71LIFNGAGSDD816 pKa = 3.53TATGGDD822 pKa = 4.1GDD824 pKa = 3.95DD825 pKa = 4.15TLWGGGGDD833 pKa = 4.14DD834 pKa = 5.25LLTGGTGADD843 pKa = 2.65IFAFTSTNGSDD854 pKa = 2.98TVTDD858 pKa = 4.08FDD860 pKa = 3.91AAEE863 pKa = 5.51DD864 pKa = 3.59ILLLDD869 pKa = 4.17GSGFADD875 pKa = 4.74LAALKK880 pKa = 10.44AATTEE885 pKa = 4.25DD886 pKa = 3.82GAGVTISFGDD896 pKa = 3.55TVIILSGISLDD907 pKa = 4.71DD908 pKa = 3.38LTAANVMLL916 pKa = 4.98
Molecular weight: 94.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G7EK86|A0A1G7EK86_9PROT DNA gyrase subunit B OS=Kordiimonas lacus OX=637679 GN=gyrB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLVRR12 pKa = 11.84KK13 pKa = 9.31RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.71ATVGGRR28 pKa = 11.84AVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLVRR12 pKa = 11.84KK13 pKa = 9.31RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.5GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.71ATVGGRR28 pKa = 11.84AVLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.77GRR39 pKa = 11.84KK40 pKa = 8.91RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205558 |
36 |
2644 |
330.0 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.912 ± 0.051 | 0.831 ± 0.014 |
6.447 ± 0.045 | 6.217 ± 0.042 |
4.073 ± 0.029 | 8.416 ± 0.051 |
2.104 ± 0.021 | 5.299 ± 0.03 |
4.403 ± 0.036 | 9.751 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.85 ± 0.02 | 3.015 ± 0.027 |
4.537 ± 0.028 | 3.201 ± 0.022 |
5.809 ± 0.042 | 5.498 ± 0.038 |
5.5 ± 0.032 | 7.2 ± 0.031 |
1.303 ± 0.016 | 2.632 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
