
Zhihengliuella halotolerans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Zhihengliuella
Average proteome isoelectric point is 5.97
Get precalculated fractions of proteins
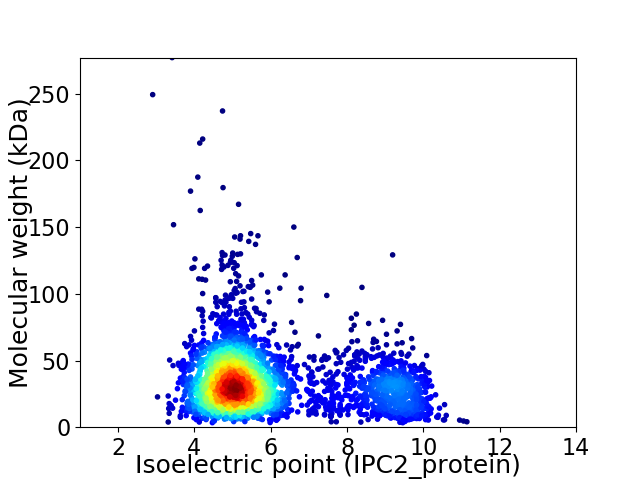
Virtual 2D-PAGE plot for 3241 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q8AAY5|A0A4Q8AAY5_9MICC Amino acid ABC transporter ATP-binding protein (PAAT family) OS=Zhihengliuella halotolerans OX=370736 GN=EV380_0213 PE=4 SV=1
MM1 pKa = 6.94TRR3 pKa = 11.84KK4 pKa = 8.3STRR7 pKa = 11.84AVGLVASLAVAAGALAGCGQSGDD30 pKa = 4.03PDD32 pKa = 3.73SLTFMFRR39 pKa = 11.84GSPDD43 pKa = 2.9EE44 pKa = 3.95EE45 pKa = 4.0AAYY48 pKa = 9.47TEE50 pKa = 4.71AIEE53 pKa = 4.57AFEE56 pKa = 4.58AEE58 pKa = 4.43SGIEE62 pKa = 3.59VDD64 pKa = 5.1MIMTTADD71 pKa = 3.14EE72 pKa = 4.55YY73 pKa = 10.5ATKK76 pKa = 10.54LRR78 pKa = 11.84AAIVGGQIPDD88 pKa = 3.16VFYY91 pKa = 10.74FDD93 pKa = 4.31PGSVEE98 pKa = 4.35SYY100 pKa = 11.17ANAGVIQEE108 pKa = 3.84ITHH111 pKa = 6.21YY112 pKa = 10.46IEE114 pKa = 5.23ASDD117 pKa = 5.41VVDD120 pKa = 5.59LDD122 pKa = 4.09NMWDD126 pKa = 3.71YY127 pKa = 11.81GVDD130 pKa = 3.15SYY132 pKa = 11.73RR133 pKa = 11.84YY134 pKa = 9.99DD135 pKa = 3.61GEE137 pKa = 4.14QLGRR141 pKa = 11.84GPLYY145 pKa = 10.56ALPKK149 pKa = 10.41DD150 pKa = 3.59VGPFSFGYY158 pKa = 9.07NATMLEE164 pKa = 4.13EE165 pKa = 5.09AGIEE169 pKa = 4.2LPDD172 pKa = 4.38PDD174 pKa = 5.53EE175 pKa = 4.96PYY177 pKa = 9.73TWEE180 pKa = 3.7EE181 pKa = 3.47WLEE184 pKa = 3.78ILEE187 pKa = 4.81EE188 pKa = 4.25VTQDD192 pKa = 2.9TDD194 pKa = 3.8GDD196 pKa = 4.09GKK198 pKa = 9.17TDD200 pKa = 3.09QWGTGLNVTWNLQAFAWSNGGEE222 pKa = 4.27WLNEE226 pKa = 3.98DD227 pKa = 3.56ATQVTVDD234 pKa = 3.42TPEE237 pKa = 3.76FAEE240 pKa = 4.09ALQYY244 pKa = 10.94FSDD247 pKa = 4.35LDD249 pKa = 3.61TEE251 pKa = 4.58YY252 pKa = 11.41GVTPSIAEE260 pKa = 3.99AQTLDD265 pKa = 3.23TYY267 pKa = 9.55QRR269 pKa = 11.84WMQGQIAFFPIAPWDD284 pKa = 3.43IPVYY288 pKa = 10.38QDD290 pKa = 6.21LDD292 pKa = 4.22FEE294 pKa = 4.91WDD296 pKa = 3.51LMPYY300 pKa = 9.62PVGRR304 pKa = 11.84TGEE307 pKa = 4.0PASWIGTLGIGVSAATANPQAAADD331 pKa = 4.16LVAHH335 pKa = 6.74LSADD339 pKa = 3.61PDD341 pKa = 4.08TQRR344 pKa = 11.84SLAEE348 pKa = 4.3AGVQIPNLIDD358 pKa = 3.34MAHH361 pKa = 6.72EE362 pKa = 4.36FAADD366 pKa = 3.73DD367 pKa = 3.65STPPVNKK374 pKa = 9.92QEE376 pKa = 4.02FLDD379 pKa = 4.26VIEE382 pKa = 5.33DD383 pKa = 3.62YY384 pKa = 11.4GRR386 pKa = 11.84ALPPTYY392 pKa = 9.75TYY394 pKa = 11.1NPQWYY399 pKa = 9.55DD400 pKa = 3.38EE401 pKa = 5.18LYY403 pKa = 10.87TNIQPVLEE411 pKa = 4.29GRR413 pKa = 11.84KK414 pKa = 7.13TAADD418 pKa = 3.97YY419 pKa = 10.73LAEE422 pKa = 4.34AQPRR426 pKa = 11.84MQAFLDD432 pKa = 3.7MANDD436 pKa = 3.45QSEE439 pKa = 4.37MSRR442 pKa = 11.84GTKK445 pKa = 9.6
MM1 pKa = 6.94TRR3 pKa = 11.84KK4 pKa = 8.3STRR7 pKa = 11.84AVGLVASLAVAAGALAGCGQSGDD30 pKa = 4.03PDD32 pKa = 3.73SLTFMFRR39 pKa = 11.84GSPDD43 pKa = 2.9EE44 pKa = 3.95EE45 pKa = 4.0AAYY48 pKa = 9.47TEE50 pKa = 4.71AIEE53 pKa = 4.57AFEE56 pKa = 4.58AEE58 pKa = 4.43SGIEE62 pKa = 3.59VDD64 pKa = 5.1MIMTTADD71 pKa = 3.14EE72 pKa = 4.55YY73 pKa = 10.5ATKK76 pKa = 10.54LRR78 pKa = 11.84AAIVGGQIPDD88 pKa = 3.16VFYY91 pKa = 10.74FDD93 pKa = 4.31PGSVEE98 pKa = 4.35SYY100 pKa = 11.17ANAGVIQEE108 pKa = 3.84ITHH111 pKa = 6.21YY112 pKa = 10.46IEE114 pKa = 5.23ASDD117 pKa = 5.41VVDD120 pKa = 5.59LDD122 pKa = 4.09NMWDD126 pKa = 3.71YY127 pKa = 11.81GVDD130 pKa = 3.15SYY132 pKa = 11.73RR133 pKa = 11.84YY134 pKa = 9.99DD135 pKa = 3.61GEE137 pKa = 4.14QLGRR141 pKa = 11.84GPLYY145 pKa = 10.56ALPKK149 pKa = 10.41DD150 pKa = 3.59VGPFSFGYY158 pKa = 9.07NATMLEE164 pKa = 4.13EE165 pKa = 5.09AGIEE169 pKa = 4.2LPDD172 pKa = 4.38PDD174 pKa = 5.53EE175 pKa = 4.96PYY177 pKa = 9.73TWEE180 pKa = 3.7EE181 pKa = 3.47WLEE184 pKa = 3.78ILEE187 pKa = 4.81EE188 pKa = 4.25VTQDD192 pKa = 2.9TDD194 pKa = 3.8GDD196 pKa = 4.09GKK198 pKa = 9.17TDD200 pKa = 3.09QWGTGLNVTWNLQAFAWSNGGEE222 pKa = 4.27WLNEE226 pKa = 3.98DD227 pKa = 3.56ATQVTVDD234 pKa = 3.42TPEE237 pKa = 3.76FAEE240 pKa = 4.09ALQYY244 pKa = 10.94FSDD247 pKa = 4.35LDD249 pKa = 3.61TEE251 pKa = 4.58YY252 pKa = 11.41GVTPSIAEE260 pKa = 3.99AQTLDD265 pKa = 3.23TYY267 pKa = 9.55QRR269 pKa = 11.84WMQGQIAFFPIAPWDD284 pKa = 3.43IPVYY288 pKa = 10.38QDD290 pKa = 6.21LDD292 pKa = 4.22FEE294 pKa = 4.91WDD296 pKa = 3.51LMPYY300 pKa = 9.62PVGRR304 pKa = 11.84TGEE307 pKa = 4.0PASWIGTLGIGVSAATANPQAAADD331 pKa = 4.16LVAHH335 pKa = 6.74LSADD339 pKa = 3.61PDD341 pKa = 4.08TQRR344 pKa = 11.84SLAEE348 pKa = 4.3AGVQIPNLIDD358 pKa = 3.34MAHH361 pKa = 6.72EE362 pKa = 4.36FAADD366 pKa = 3.73DD367 pKa = 3.65STPPVNKK374 pKa = 9.92QEE376 pKa = 4.02FLDD379 pKa = 4.26VIEE382 pKa = 5.33DD383 pKa = 3.62YY384 pKa = 11.4GRR386 pKa = 11.84ALPPTYY392 pKa = 9.75TYY394 pKa = 11.1NPQWYY399 pKa = 9.55DD400 pKa = 3.38EE401 pKa = 5.18LYY403 pKa = 10.87TNIQPVLEE411 pKa = 4.29GRR413 pKa = 11.84KK414 pKa = 7.13TAADD418 pKa = 3.97YY419 pKa = 10.73LAEE422 pKa = 4.34AQPRR426 pKa = 11.84MQAFLDD432 pKa = 3.7MANDD436 pKa = 3.45QSEE439 pKa = 4.37MSRR442 pKa = 11.84GTKK445 pKa = 9.6
Molecular weight: 48.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q8AF36|A0A4Q8AF36_9MICC DNA-binding protein HU-beta OS=Zhihengliuella halotolerans OX=370736 GN=EV380_2057 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1082384 |
31 |
2601 |
334.0 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.549 ± 0.059 | 0.595 ± 0.01 |
6.098 ± 0.042 | 6.209 ± 0.043 |
3.203 ± 0.03 | 9.138 ± 0.04 |
2.102 ± 0.018 | 4.199 ± 0.036 |
2.234 ± 0.033 | 9.992 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.957 ± 0.018 | 2.231 ± 0.027 |
5.259 ± 0.032 | 2.794 ± 0.023 |
6.893 ± 0.051 | 5.606 ± 0.033 |
5.846 ± 0.034 | 8.582 ± 0.041 |
1.469 ± 0.018 | 2.047 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
