
Haloterrigena daqingensis
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Haloterrigena
Average proteome isoelectric point is 4.75
Get precalculated fractions of proteins
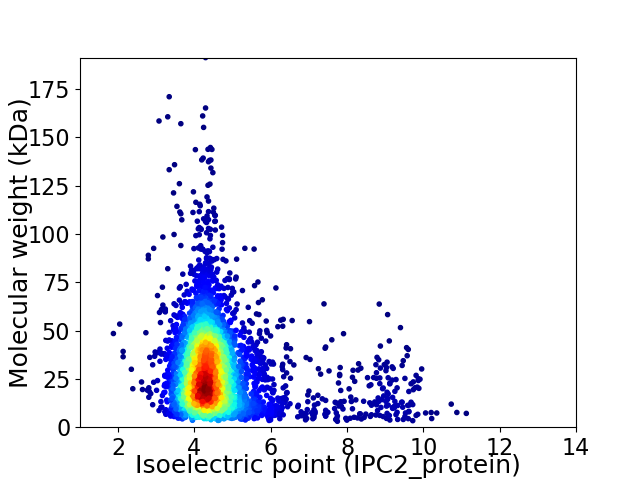
Virtual 2D-PAGE plot for 3795 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
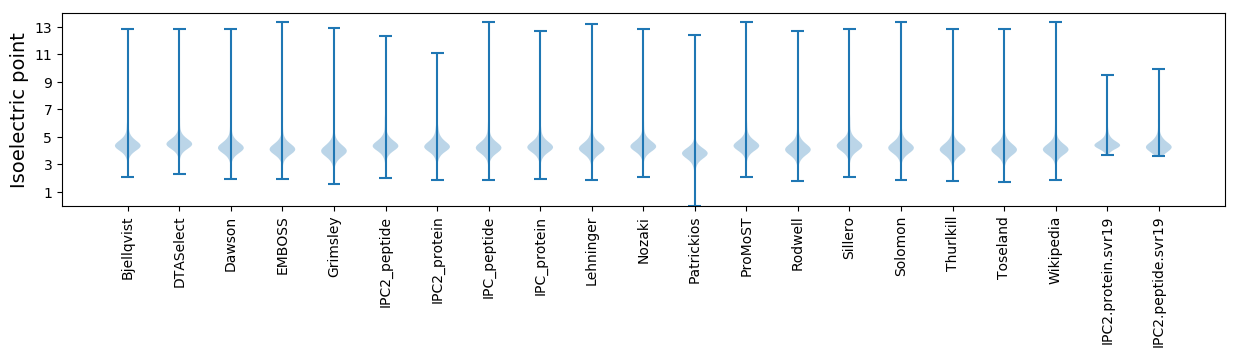
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1N6X400|A0A1N6X400_9EURY Histidinol dehydrogenase OS=Haloterrigena daqingensis OX=588898 GN=hisD PE=3 SV=1
MM1 pKa = 7.2VSYY4 pKa = 10.96SPTLLEE10 pKa = 3.72EE11 pKa = 4.62HH12 pKa = 6.47GEE14 pKa = 3.81KK15 pKa = 10.26RR16 pKa = 11.84AIINFDD22 pKa = 3.86RR23 pKa = 11.84PDD25 pKa = 3.45EE26 pKa = 4.22YY27 pKa = 11.41YY28 pKa = 10.85GPCEE32 pKa = 3.92TRR34 pKa = 11.84SEE36 pKa = 4.32KK37 pKa = 10.22IAEE40 pKa = 4.2LKK42 pKa = 9.73EE43 pKa = 4.53CCNSTHH49 pKa = 7.07APLKK53 pKa = 10.8DD54 pKa = 4.7FISSTAGIEE63 pKa = 4.01LLSDD67 pKa = 3.47HH68 pKa = 6.78WAGSTATVRR77 pKa = 11.84IDD79 pKa = 3.74FNTFDD84 pKa = 3.89VDD86 pKa = 3.83DD87 pKa = 5.68LEE89 pKa = 5.66DD90 pKa = 3.29VDD92 pKa = 4.35YY93 pKa = 11.18VKK95 pKa = 10.91SVVVDD100 pKa = 4.06DD101 pKa = 4.22GTLKK105 pKa = 10.69INSDD109 pKa = 4.25DD110 pKa = 4.24GADD113 pKa = 3.36TYY115 pKa = 11.77LPGDD119 pKa = 3.75VDD121 pKa = 3.33WSLEE125 pKa = 3.79QTQVHH130 pKa = 5.65EE131 pKa = 4.49FWTEE135 pKa = 3.48YY136 pKa = 10.15GARR139 pKa = 11.84GEE141 pKa = 4.19NVKK144 pKa = 10.61VAVTDD149 pKa = 3.42TGIDD153 pKa = 3.61PDD155 pKa = 4.07HH156 pKa = 6.96EE157 pKa = 4.32ALEE160 pKa = 5.29LYY162 pKa = 10.22TDD164 pKa = 4.41DD165 pKa = 5.86PDD167 pKa = 6.24DD168 pKa = 3.55EE169 pKa = 4.88TYY171 pKa = 10.51PGGWAEE177 pKa = 5.01FDD179 pKa = 3.62LDD181 pKa = 3.85GNEE184 pKa = 4.8IEE186 pKa = 6.14DD187 pKa = 3.86SEE189 pKa = 4.1PWYY192 pKa = 10.24DD193 pKa = 3.59FWHH196 pKa = 6.79GIEE199 pKa = 3.89VSGPIKK205 pKa = 10.51GKK207 pKa = 9.53PPEE210 pKa = 4.48SADD213 pKa = 3.51EE214 pKa = 4.51EE215 pKa = 5.11YY216 pKa = 10.41MGVAPEE222 pKa = 4.58CDD224 pKa = 2.9LMMVKK229 pKa = 9.94MNDD232 pKa = 3.37PDD234 pKa = 3.74EE235 pKa = 4.71TDD237 pKa = 3.5TTTHH241 pKa = 5.91SAMIAALEE249 pKa = 4.02WCLEE253 pKa = 3.99NGADD257 pKa = 4.94IINSSWGNYY266 pKa = 7.84ARR268 pKa = 11.84QSPPYY273 pKa = 9.84EE274 pKa = 4.66LDD276 pKa = 2.96LYY278 pKa = 11.67YY279 pKa = 10.63DD280 pKa = 4.13IFQDD284 pKa = 4.25ALDD287 pKa = 4.04MNCISVGSSGNTDD300 pKa = 2.98EE301 pKa = 5.14GGEE304 pKa = 4.03IGPAPGSYY312 pKa = 10.79YY313 pKa = 10.74NAISVGGVEE322 pKa = 5.1EE323 pKa = 4.17SGEE326 pKa = 4.35MYY328 pKa = 10.66GSSTGRR334 pKa = 11.84TIEE337 pKa = 3.84EE338 pKa = 4.48SDD340 pKa = 2.76WDD342 pKa = 4.26EE343 pKa = 5.95YY344 pKa = 10.23PDD346 pKa = 3.8EE347 pKa = 4.77WPDD350 pKa = 3.33EE351 pKa = 4.19YY352 pKa = 10.53IIPDD356 pKa = 3.36ITAGGRR362 pKa = 11.84NVPTTTEE369 pKa = 3.84DD370 pKa = 3.6DD371 pKa = 4.23EE372 pKa = 5.01YY373 pKa = 10.46GTASGTSFAAPIVTGHH389 pKa = 6.62IALLLSYY396 pKa = 10.82SRR398 pKa = 11.84TQTEE402 pKa = 3.59SDD404 pKa = 3.32ADD406 pKa = 3.59INDD409 pKa = 4.2IISALKK415 pKa = 8.03GTAWKK420 pKa = 9.86PDD422 pKa = 3.3DD423 pKa = 3.93WSDD426 pKa = 4.52DD427 pKa = 3.71DD428 pKa = 6.14FEE430 pKa = 7.32DD431 pKa = 4.78GSEE434 pKa = 3.98EE435 pKa = 4.24AGEE438 pKa = 3.98HH439 pKa = 5.26DD440 pKa = 3.37TRR442 pKa = 11.84YY443 pKa = 10.24GCGIAKK449 pKa = 10.13VLDD452 pKa = 3.98AQEE455 pKa = 4.53KK456 pKa = 8.99LRR458 pKa = 11.84DD459 pKa = 3.58MRR461 pKa = 11.84SITGEE466 pKa = 4.08VIFNSIPTNDD476 pKa = 3.26AKK478 pKa = 11.36VYY480 pKa = 10.85VIDD483 pKa = 4.98RR484 pKa = 11.84NSNDD488 pKa = 3.19LAGVLEE494 pKa = 4.28PSEE497 pKa = 4.64GEE499 pKa = 3.69FRR501 pKa = 11.84YY502 pKa = 10.43EE503 pKa = 4.41GGSSDD508 pKa = 3.87DD509 pKa = 3.9SYY511 pKa = 11.56HH512 pKa = 7.25IGVVADD518 pKa = 4.68DD519 pKa = 4.29DD520 pKa = 4.03
MM1 pKa = 7.2VSYY4 pKa = 10.96SPTLLEE10 pKa = 3.72EE11 pKa = 4.62HH12 pKa = 6.47GEE14 pKa = 3.81KK15 pKa = 10.26RR16 pKa = 11.84AIINFDD22 pKa = 3.86RR23 pKa = 11.84PDD25 pKa = 3.45EE26 pKa = 4.22YY27 pKa = 11.41YY28 pKa = 10.85GPCEE32 pKa = 3.92TRR34 pKa = 11.84SEE36 pKa = 4.32KK37 pKa = 10.22IAEE40 pKa = 4.2LKK42 pKa = 9.73EE43 pKa = 4.53CCNSTHH49 pKa = 7.07APLKK53 pKa = 10.8DD54 pKa = 4.7FISSTAGIEE63 pKa = 4.01LLSDD67 pKa = 3.47HH68 pKa = 6.78WAGSTATVRR77 pKa = 11.84IDD79 pKa = 3.74FNTFDD84 pKa = 3.89VDD86 pKa = 3.83DD87 pKa = 5.68LEE89 pKa = 5.66DD90 pKa = 3.29VDD92 pKa = 4.35YY93 pKa = 11.18VKK95 pKa = 10.91SVVVDD100 pKa = 4.06DD101 pKa = 4.22GTLKK105 pKa = 10.69INSDD109 pKa = 4.25DD110 pKa = 4.24GADD113 pKa = 3.36TYY115 pKa = 11.77LPGDD119 pKa = 3.75VDD121 pKa = 3.33WSLEE125 pKa = 3.79QTQVHH130 pKa = 5.65EE131 pKa = 4.49FWTEE135 pKa = 3.48YY136 pKa = 10.15GARR139 pKa = 11.84GEE141 pKa = 4.19NVKK144 pKa = 10.61VAVTDD149 pKa = 3.42TGIDD153 pKa = 3.61PDD155 pKa = 4.07HH156 pKa = 6.96EE157 pKa = 4.32ALEE160 pKa = 5.29LYY162 pKa = 10.22TDD164 pKa = 4.41DD165 pKa = 5.86PDD167 pKa = 6.24DD168 pKa = 3.55EE169 pKa = 4.88TYY171 pKa = 10.51PGGWAEE177 pKa = 5.01FDD179 pKa = 3.62LDD181 pKa = 3.85GNEE184 pKa = 4.8IEE186 pKa = 6.14DD187 pKa = 3.86SEE189 pKa = 4.1PWYY192 pKa = 10.24DD193 pKa = 3.59FWHH196 pKa = 6.79GIEE199 pKa = 3.89VSGPIKK205 pKa = 10.51GKK207 pKa = 9.53PPEE210 pKa = 4.48SADD213 pKa = 3.51EE214 pKa = 4.51EE215 pKa = 5.11YY216 pKa = 10.41MGVAPEE222 pKa = 4.58CDD224 pKa = 2.9LMMVKK229 pKa = 9.94MNDD232 pKa = 3.37PDD234 pKa = 3.74EE235 pKa = 4.71TDD237 pKa = 3.5TTTHH241 pKa = 5.91SAMIAALEE249 pKa = 4.02WCLEE253 pKa = 3.99NGADD257 pKa = 4.94IINSSWGNYY266 pKa = 7.84ARR268 pKa = 11.84QSPPYY273 pKa = 9.84EE274 pKa = 4.66LDD276 pKa = 2.96LYY278 pKa = 11.67YY279 pKa = 10.63DD280 pKa = 4.13IFQDD284 pKa = 4.25ALDD287 pKa = 4.04MNCISVGSSGNTDD300 pKa = 2.98EE301 pKa = 5.14GGEE304 pKa = 4.03IGPAPGSYY312 pKa = 10.79YY313 pKa = 10.74NAISVGGVEE322 pKa = 5.1EE323 pKa = 4.17SGEE326 pKa = 4.35MYY328 pKa = 10.66GSSTGRR334 pKa = 11.84TIEE337 pKa = 3.84EE338 pKa = 4.48SDD340 pKa = 2.76WDD342 pKa = 4.26EE343 pKa = 5.95YY344 pKa = 10.23PDD346 pKa = 3.8EE347 pKa = 4.77WPDD350 pKa = 3.33EE351 pKa = 4.19YY352 pKa = 10.53IIPDD356 pKa = 3.36ITAGGRR362 pKa = 11.84NVPTTTEE369 pKa = 3.84DD370 pKa = 3.6DD371 pKa = 4.23EE372 pKa = 5.01YY373 pKa = 10.46GTASGTSFAAPIVTGHH389 pKa = 6.62IALLLSYY396 pKa = 10.82SRR398 pKa = 11.84TQTEE402 pKa = 3.59SDD404 pKa = 3.32ADD406 pKa = 3.59INDD409 pKa = 4.2IISALKK415 pKa = 8.03GTAWKK420 pKa = 9.86PDD422 pKa = 3.3DD423 pKa = 3.93WSDD426 pKa = 4.52DD427 pKa = 3.71DD428 pKa = 6.14FEE430 pKa = 7.32DD431 pKa = 4.78GSEE434 pKa = 3.98EE435 pKa = 4.24AGEE438 pKa = 3.98HH439 pKa = 5.26DD440 pKa = 3.37TRR442 pKa = 11.84YY443 pKa = 10.24GCGIAKK449 pKa = 10.13VLDD452 pKa = 3.98AQEE455 pKa = 4.53KK456 pKa = 8.99LRR458 pKa = 11.84DD459 pKa = 3.58MRR461 pKa = 11.84SITGEE466 pKa = 4.08VIFNSIPTNDD476 pKa = 3.26AKK478 pKa = 11.36VYY480 pKa = 10.85VIDD483 pKa = 4.98RR484 pKa = 11.84NSNDD488 pKa = 3.19LAGVLEE494 pKa = 4.28PSEE497 pKa = 4.64GEE499 pKa = 3.69FRR501 pKa = 11.84YY502 pKa = 10.43EE503 pKa = 4.41GGSSDD508 pKa = 3.87DD509 pKa = 3.9SYY511 pKa = 11.56HH512 pKa = 7.25IGVVADD518 pKa = 4.68DD519 pKa = 4.29DD520 pKa = 4.03
Molecular weight: 57.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1N7FFX8|A0A1N7FFX8_9EURY Uncharacterized protein OS=Haloterrigena daqingensis OX=588898 GN=BB347_17005 PE=4 SV=1
MM1 pKa = 7.43GGVAGLSLVGFSGVASAGKK20 pKa = 9.98NGRR23 pKa = 11.84NGKK26 pKa = 9.08NGKK29 pKa = 8.78NGKK32 pKa = 9.45NNGRR36 pKa = 11.84NGKK39 pKa = 9.54HH40 pKa = 5.94NGRR43 pKa = 11.84NGKK46 pKa = 9.08NGNGRR51 pKa = 11.84NGKK54 pKa = 9.89RR55 pKa = 11.84NGRR58 pKa = 11.84NGKK61 pKa = 9.86RR62 pKa = 11.84NGRR65 pKa = 11.84NGKK68 pKa = 9.19NGNN71 pKa = 3.54
MM1 pKa = 7.43GGVAGLSLVGFSGVASAGKK20 pKa = 9.98NGRR23 pKa = 11.84NGKK26 pKa = 9.08NGKK29 pKa = 8.78NGKK32 pKa = 9.45NNGRR36 pKa = 11.84NGKK39 pKa = 9.54HH40 pKa = 5.94NGRR43 pKa = 11.84NGKK46 pKa = 9.08NGNGRR51 pKa = 11.84NGKK54 pKa = 9.89RR55 pKa = 11.84NGRR58 pKa = 11.84NGKK61 pKa = 9.86RR62 pKa = 11.84NGRR65 pKa = 11.84NGKK68 pKa = 9.19NGNN71 pKa = 3.54
Molecular weight: 7.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1090797 |
29 |
1729 |
287.4 |
31.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.948 ± 0.055 | 0.764 ± 0.014 |
8.759 ± 0.074 | 9.505 ± 0.062 |
3.302 ± 0.03 | 8.092 ± 0.039 |
2.115 ± 0.02 | 4.595 ± 0.036 |
1.804 ± 0.028 | 8.851 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.78 ± 0.018 | 2.426 ± 0.024 |
4.491 ± 0.025 | 2.587 ± 0.025 |
6.039 ± 0.039 | 5.997 ± 0.034 |
6.561 ± 0.03 | 8.501 ± 0.041 |
1.129 ± 0.016 | 2.754 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
