
Gracilibacillus ureilyticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Gracilibacillus
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
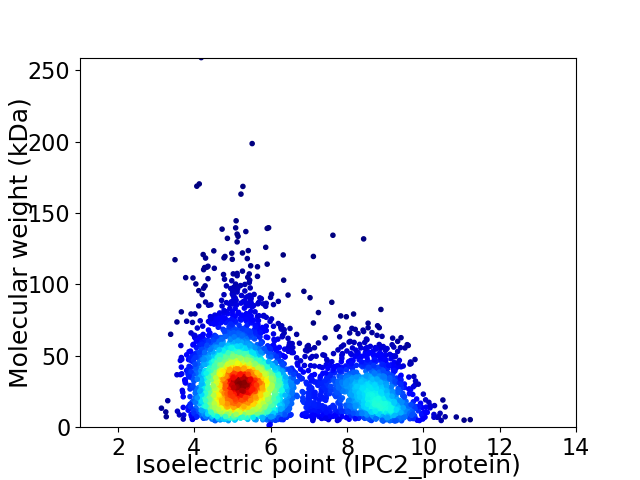
Virtual 2D-PAGE plot for 3985 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9MYV4|A0A1H9MYV4_9BACI PadR family transcriptional regulator regulatory protein PadR OS=Gracilibacillus ureilyticus OX=531814 GN=SAMN04487944_102232 PE=4 SV=1
MM1 pKa = 7.51RR2 pKa = 11.84KK3 pKa = 9.65SYY5 pKa = 11.3ALLVMLLSAMLVLSACAGGGSSSGDD30 pKa = 3.05KK31 pKa = 11.29GEE33 pKa = 4.13AAEE36 pKa = 5.49DD37 pKa = 3.82GTKK40 pKa = 10.57EE41 pKa = 3.77LTFMFRR47 pKa = 11.84GGTAEE52 pKa = 3.76EE53 pKa = 3.94EE54 pKa = 4.56AYY56 pKa = 10.55QKK58 pKa = 11.1AIDD61 pKa = 3.88QFEE64 pKa = 4.3ADD66 pKa = 3.61NEE68 pKa = 4.54GVKK71 pKa = 10.96VNMIVTAADD80 pKa = 3.39QYY82 pKa = 9.23STKK85 pKa = 10.52LKK87 pKa = 10.75AAITGNQVPDD97 pKa = 3.26VFYY100 pKa = 10.64IEE102 pKa = 4.66PGQLRR107 pKa = 11.84SYY109 pKa = 9.16VHH111 pKa = 6.79SNILLDD117 pKa = 3.43LTEE120 pKa = 4.56YY121 pKa = 10.61IEE123 pKa = 4.81NNDD126 pKa = 4.79NINLDD131 pKa = 4.24NIWQYY136 pKa = 11.69GVDD139 pKa = 3.35SYY141 pKa = 11.82RR142 pKa = 11.84FDD144 pKa = 5.54GEE146 pKa = 4.45TQGQGAIYY154 pKa = 10.49AMPKK158 pKa = 10.31DD159 pKa = 3.77VGPFSLGYY167 pKa = 9.33NKK169 pKa = 10.05TMFEE173 pKa = 4.13EE174 pKa = 5.05AGLEE178 pKa = 4.07LPDD181 pKa = 4.39EE182 pKa = 4.83DD183 pKa = 6.2VPMTWDD189 pKa = 3.32EE190 pKa = 4.64FIEE193 pKa = 4.32ANQALTKK200 pKa = 10.3DD201 pKa = 3.35TDD203 pKa = 3.71GDD205 pKa = 4.31GEE207 pKa = 4.07IDD209 pKa = 3.62QYY211 pKa = 11.44GTGFNVNWVLQSFVWSNGGSFIDD234 pKa = 3.67EE235 pKa = 4.43TQTKK239 pKa = 7.54VTVDD243 pKa = 3.26TPEE246 pKa = 3.9FAEE249 pKa = 4.16ALQWFADD256 pKa = 3.64HH257 pKa = 6.3QNVYY261 pKa = 10.19EE262 pKa = 4.08ITPSIEE268 pKa = 3.9DD269 pKa = 3.57AQTLDD274 pKa = 3.24TYY276 pKa = 9.51QRR278 pKa = 11.84WMQGQLAFFPVAPWDD293 pKa = 3.5LATFDD298 pKa = 3.97TEE300 pKa = 5.61LDD302 pKa = 3.48FEE304 pKa = 5.08YY305 pKa = 11.11DD306 pKa = 3.93VIPYY310 pKa = 8.78PAGKK314 pKa = 8.25TGEE317 pKa = 4.16SATWIGTLGIGVAQSSEE334 pKa = 4.13YY335 pKa = 10.25PEE337 pKa = 4.58LATEE341 pKa = 4.21LVYY344 pKa = 11.22YY345 pKa = 8.54LTASEE350 pKa = 4.84EE351 pKa = 4.39GQQALVDD358 pKa = 3.86ARR360 pKa = 11.84IQIPNLIDD368 pKa = 3.53MAQEE372 pKa = 3.77WAADD376 pKa = 3.65EE377 pKa = 4.72SMAPANKK384 pKa = 9.86EE385 pKa = 3.62EE386 pKa = 3.86YY387 pKa = 10.32LQIIEE392 pKa = 4.57DD393 pKa = 3.81YY394 pKa = 11.21GRR396 pKa = 11.84ALPANNTYY404 pKa = 9.78NAEE407 pKa = 3.86WYY409 pKa = 9.87DD410 pKa = 3.75YY411 pKa = 10.93FFTNVQPVLDD421 pKa = 4.12GDD423 pKa = 3.7ITAAEE428 pKa = 4.48YY429 pKa = 10.98VEE431 pKa = 4.39QVQPKK436 pKa = 6.83MQEE439 pKa = 3.95YY440 pKa = 10.31LDD442 pKa = 3.67NARR445 pKa = 11.84EE446 pKa = 4.07LEE448 pKa = 4.29EE449 pKa = 4.34QSKK452 pKa = 8.56QQ453 pKa = 3.31
MM1 pKa = 7.51RR2 pKa = 11.84KK3 pKa = 9.65SYY5 pKa = 11.3ALLVMLLSAMLVLSACAGGGSSSGDD30 pKa = 3.05KK31 pKa = 11.29GEE33 pKa = 4.13AAEE36 pKa = 5.49DD37 pKa = 3.82GTKK40 pKa = 10.57EE41 pKa = 3.77LTFMFRR47 pKa = 11.84GGTAEE52 pKa = 3.76EE53 pKa = 3.94EE54 pKa = 4.56AYY56 pKa = 10.55QKK58 pKa = 11.1AIDD61 pKa = 3.88QFEE64 pKa = 4.3ADD66 pKa = 3.61NEE68 pKa = 4.54GVKK71 pKa = 10.96VNMIVTAADD80 pKa = 3.39QYY82 pKa = 9.23STKK85 pKa = 10.52LKK87 pKa = 10.75AAITGNQVPDD97 pKa = 3.26VFYY100 pKa = 10.64IEE102 pKa = 4.66PGQLRR107 pKa = 11.84SYY109 pKa = 9.16VHH111 pKa = 6.79SNILLDD117 pKa = 3.43LTEE120 pKa = 4.56YY121 pKa = 10.61IEE123 pKa = 4.81NNDD126 pKa = 4.79NINLDD131 pKa = 4.24NIWQYY136 pKa = 11.69GVDD139 pKa = 3.35SYY141 pKa = 11.82RR142 pKa = 11.84FDD144 pKa = 5.54GEE146 pKa = 4.45TQGQGAIYY154 pKa = 10.49AMPKK158 pKa = 10.31DD159 pKa = 3.77VGPFSLGYY167 pKa = 9.33NKK169 pKa = 10.05TMFEE173 pKa = 4.13EE174 pKa = 5.05AGLEE178 pKa = 4.07LPDD181 pKa = 4.39EE182 pKa = 4.83DD183 pKa = 6.2VPMTWDD189 pKa = 3.32EE190 pKa = 4.64FIEE193 pKa = 4.32ANQALTKK200 pKa = 10.3DD201 pKa = 3.35TDD203 pKa = 3.71GDD205 pKa = 4.31GEE207 pKa = 4.07IDD209 pKa = 3.62QYY211 pKa = 11.44GTGFNVNWVLQSFVWSNGGSFIDD234 pKa = 3.67EE235 pKa = 4.43TQTKK239 pKa = 7.54VTVDD243 pKa = 3.26TPEE246 pKa = 3.9FAEE249 pKa = 4.16ALQWFADD256 pKa = 3.64HH257 pKa = 6.3QNVYY261 pKa = 10.19EE262 pKa = 4.08ITPSIEE268 pKa = 3.9DD269 pKa = 3.57AQTLDD274 pKa = 3.24TYY276 pKa = 9.51QRR278 pKa = 11.84WMQGQLAFFPVAPWDD293 pKa = 3.5LATFDD298 pKa = 3.97TEE300 pKa = 5.61LDD302 pKa = 3.48FEE304 pKa = 5.08YY305 pKa = 11.11DD306 pKa = 3.93VIPYY310 pKa = 8.78PAGKK314 pKa = 8.25TGEE317 pKa = 4.16SATWIGTLGIGVAQSSEE334 pKa = 4.13YY335 pKa = 10.25PEE337 pKa = 4.58LATEE341 pKa = 4.21LVYY344 pKa = 11.22YY345 pKa = 8.54LTASEE350 pKa = 4.84EE351 pKa = 4.39GQQALVDD358 pKa = 3.86ARR360 pKa = 11.84IQIPNLIDD368 pKa = 3.53MAQEE372 pKa = 3.77WAADD376 pKa = 3.65EE377 pKa = 4.72SMAPANKK384 pKa = 9.86EE385 pKa = 3.62EE386 pKa = 3.86YY387 pKa = 10.32LQIIEE392 pKa = 4.57DD393 pKa = 3.81YY394 pKa = 11.21GRR396 pKa = 11.84ALPANNTYY404 pKa = 9.78NAEE407 pKa = 3.86WYY409 pKa = 9.87DD410 pKa = 3.75YY411 pKa = 10.93FFTNVQPVLDD421 pKa = 4.12GDD423 pKa = 3.7ITAAEE428 pKa = 4.48YY429 pKa = 10.98VEE431 pKa = 4.39QVQPKK436 pKa = 6.83MQEE439 pKa = 3.95YY440 pKa = 10.31LDD442 pKa = 3.67NARR445 pKa = 11.84EE446 pKa = 4.07LEE448 pKa = 4.29EE449 pKa = 4.34QSKK452 pKa = 8.56QQ453 pKa = 3.31
Molecular weight: 50.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9V064|A0A1H9V064_9BACI tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG OS=Gracilibacillus ureilyticus OX=531814 GN=mnmG PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.75VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.23RR12 pKa = 11.84KK13 pKa = 8.22KK14 pKa = 8.69VHH16 pKa = 5.46GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTKK25 pKa = 10.19NGRR28 pKa = 11.84KK29 pKa = 8.75VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1176553 |
11 |
2311 |
295.2 |
33.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.62 ± 0.04 | 0.65 ± 0.012 |
5.403 ± 0.033 | 7.84 ± 0.052 |
4.595 ± 0.037 | 6.497 ± 0.038 |
2.098 ± 0.022 | 8.258 ± 0.044 |
6.632 ± 0.04 | 9.56 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.749 ± 0.02 | 4.812 ± 0.031 |
3.445 ± 0.021 | 3.958 ± 0.029 |
3.808 ± 0.029 | 6.024 ± 0.03 |
5.432 ± 0.024 | 6.749 ± 0.033 |
1.119 ± 0.017 | 3.752 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
