
Cladosporium cladosporioides virus 1
Taxonomy: Viruses; Riboviria; Polymycoviridae; Polymycovirus; Cladosporium cladosporioides polymycovirus 1
Average proteome isoelectric point is 7.41
Get precalculated fractions of proteins
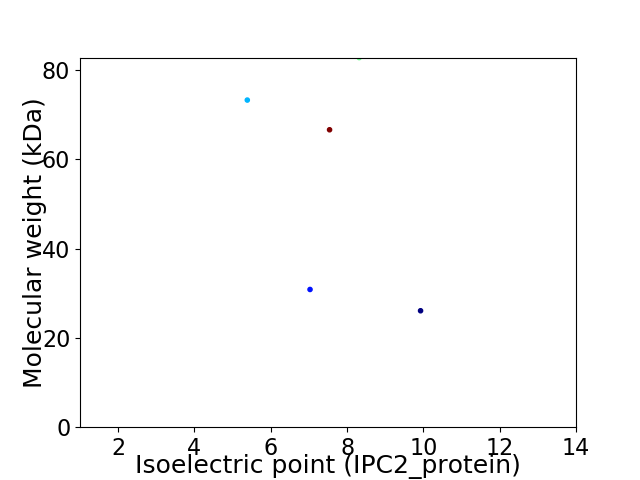
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076JWY5|A0A076JWY5_9VIRU Uncharacterized protein OS=Cladosporium cladosporioides virus 1 OX=1529605 PE=4 SV=1
MM1 pKa = 7.84ADD3 pKa = 3.94LARR6 pKa = 11.84LRR8 pKa = 11.84HH9 pKa = 5.66MLLDD13 pKa = 3.85PTEE16 pKa = 4.3SAVLLASMCHH26 pKa = 6.13HH27 pKa = 7.77LIPDD31 pKa = 3.85PTSVDD36 pKa = 3.23LFDD39 pKa = 4.51YY40 pKa = 10.3SFPPRR45 pKa = 11.84ARR47 pKa = 11.84DD48 pKa = 3.63WIISAPFPLFGPAIPASKK66 pKa = 10.64SSFEE70 pKa = 5.06AILEE74 pKa = 4.01PLRR77 pKa = 11.84AMEE80 pKa = 4.16YY81 pKa = 10.73NSLDD85 pKa = 3.26MAAVLMRR92 pKa = 11.84SMRR95 pKa = 11.84VADD98 pKa = 3.66NPFAKK103 pKa = 9.08VTRR106 pKa = 11.84GEE108 pKa = 4.29TTGDD112 pKa = 3.34VAQSRR117 pKa = 11.84AGAAQHH123 pKa = 6.7AAQSPIEE130 pKa = 3.9FAIYY134 pKa = 10.34DD135 pKa = 3.74VLRR138 pKa = 11.84DD139 pKa = 3.45AGYY142 pKa = 9.12TSVSDD147 pKa = 4.43ALSKK151 pKa = 10.73LQSFLTSTKK160 pKa = 10.77AFVHH164 pKa = 6.73PITRR168 pKa = 11.84QFSRR172 pKa = 11.84HH173 pKa = 4.65RR174 pKa = 11.84SAALFLINKK183 pKa = 8.37PGARR187 pKa = 11.84GPTAFLTPFFTSKK200 pKa = 10.59EE201 pKa = 4.07ATKK204 pKa = 10.85LFAAVVFRR212 pKa = 11.84SDD214 pKa = 2.82ILTSLSRR221 pKa = 11.84LNAARR226 pKa = 11.84GVTVLGYY233 pKa = 10.24AARR236 pKa = 11.84ASLEE240 pKa = 3.74AVRR243 pKa = 11.84RR244 pKa = 11.84IVEE247 pKa = 3.92PAHH250 pKa = 6.77AIVNEE255 pKa = 4.01YY256 pKa = 11.05SFDD259 pKa = 3.8PQTLSFIDD267 pKa = 3.86PAGDD271 pKa = 3.45PTYY274 pKa = 10.36HH275 pKa = 5.85ITDD278 pKa = 3.86RR279 pKa = 11.84TPSVVVAFGRR289 pKa = 11.84VLSGGSVDD297 pKa = 3.85ACSKK301 pKa = 10.97ASDD304 pKa = 4.02LRR306 pKa = 11.84TSLDD310 pKa = 3.33IPGGLPDD317 pKa = 3.28VARR320 pKa = 11.84EE321 pKa = 4.08YY322 pKa = 10.93FSSTGNVEE330 pKa = 3.82DD331 pKa = 4.88TIRR334 pKa = 11.84SAATALSDD342 pKa = 3.46FRR344 pKa = 11.84SFRR347 pKa = 11.84DD348 pKa = 3.41SHH350 pKa = 6.58PGGHH354 pKa = 6.32EE355 pKa = 4.03LNKK358 pKa = 10.82DD359 pKa = 3.15SGLSFFSRR367 pKa = 11.84DD368 pKa = 2.79RR369 pKa = 11.84ATRR372 pKa = 11.84DD373 pKa = 3.06RR374 pKa = 11.84NTAACITRR382 pKa = 11.84VEE384 pKa = 4.36EE385 pKa = 4.35VACLLRR391 pKa = 11.84SQGRR395 pKa = 11.84DD396 pKa = 2.88IARR399 pKa = 11.84MAFVVEE405 pKa = 4.03WGGPLNNDD413 pKa = 3.99SILAALALTGATACVDD429 pKa = 3.55VGPGAFKK436 pKa = 11.03VGDD439 pKa = 3.94SSEE442 pKa = 4.1EE443 pKa = 3.78LATEE447 pKa = 4.06NDD449 pKa = 3.01KK450 pKa = 11.33GAYY453 pKa = 8.7VCLIPRR459 pKa = 11.84ASTRR463 pKa = 11.84GLPRR467 pKa = 11.84MPIVGYY473 pKa = 10.41EE474 pKa = 4.13SGDD477 pKa = 3.8SVTQRR482 pKa = 11.84LDD484 pKa = 3.12RR485 pKa = 11.84LLSFISGPDD494 pKa = 3.09NYY496 pKa = 10.51PPVAYY501 pKa = 8.43ITGGSTTSGAAPADD515 pKa = 3.61IIGITQARR523 pKa = 11.84AALLTDD529 pKa = 3.66YY530 pKa = 10.42LASGSFFCGDD540 pKa = 3.0ILIPDD545 pKa = 4.6VCRR548 pKa = 11.84HH549 pKa = 6.14PPDD552 pKa = 3.94GTDD555 pKa = 4.85DD556 pKa = 3.75EE557 pKa = 5.39CLACANFDD565 pKa = 3.62YY566 pKa = 11.29SVFTVGDD573 pKa = 3.59VCQLDD578 pKa = 4.02GFSLVKK584 pKa = 10.14PRR586 pKa = 11.84ASYY589 pKa = 10.79AHH591 pKa = 6.52NLHH594 pKa = 7.1LSLEE598 pKa = 4.62WIHH601 pKa = 6.01GAARR605 pKa = 11.84EE606 pKa = 4.03FSEE609 pKa = 5.04SRR611 pKa = 11.84TLLALDD617 pKa = 3.78SVVATNVARR626 pKa = 11.84NADD629 pKa = 3.2WGDD632 pKa = 3.41FVPRR636 pKa = 11.84DD637 pKa = 3.72GPRR640 pKa = 11.84RR641 pKa = 11.84PHH643 pKa = 6.24HH644 pKa = 6.56PSYY647 pKa = 10.81GLKK650 pKa = 10.12VKK652 pKa = 10.64LIIEE656 pKa = 4.09DD657 pKa = 3.92VYY659 pKa = 11.49RR660 pKa = 11.84SMAEE664 pKa = 3.77GAEE667 pKa = 4.1EE668 pKa = 5.18GIGATKK674 pKa = 10.1TIVEE678 pKa = 4.44SLAA681 pKa = 3.57
MM1 pKa = 7.84ADD3 pKa = 3.94LARR6 pKa = 11.84LRR8 pKa = 11.84HH9 pKa = 5.66MLLDD13 pKa = 3.85PTEE16 pKa = 4.3SAVLLASMCHH26 pKa = 6.13HH27 pKa = 7.77LIPDD31 pKa = 3.85PTSVDD36 pKa = 3.23LFDD39 pKa = 4.51YY40 pKa = 10.3SFPPRR45 pKa = 11.84ARR47 pKa = 11.84DD48 pKa = 3.63WIISAPFPLFGPAIPASKK66 pKa = 10.64SSFEE70 pKa = 5.06AILEE74 pKa = 4.01PLRR77 pKa = 11.84AMEE80 pKa = 4.16YY81 pKa = 10.73NSLDD85 pKa = 3.26MAAVLMRR92 pKa = 11.84SMRR95 pKa = 11.84VADD98 pKa = 3.66NPFAKK103 pKa = 9.08VTRR106 pKa = 11.84GEE108 pKa = 4.29TTGDD112 pKa = 3.34VAQSRR117 pKa = 11.84AGAAQHH123 pKa = 6.7AAQSPIEE130 pKa = 3.9FAIYY134 pKa = 10.34DD135 pKa = 3.74VLRR138 pKa = 11.84DD139 pKa = 3.45AGYY142 pKa = 9.12TSVSDD147 pKa = 4.43ALSKK151 pKa = 10.73LQSFLTSTKK160 pKa = 10.77AFVHH164 pKa = 6.73PITRR168 pKa = 11.84QFSRR172 pKa = 11.84HH173 pKa = 4.65RR174 pKa = 11.84SAALFLINKK183 pKa = 8.37PGARR187 pKa = 11.84GPTAFLTPFFTSKK200 pKa = 10.59EE201 pKa = 4.07ATKK204 pKa = 10.85LFAAVVFRR212 pKa = 11.84SDD214 pKa = 2.82ILTSLSRR221 pKa = 11.84LNAARR226 pKa = 11.84GVTVLGYY233 pKa = 10.24AARR236 pKa = 11.84ASLEE240 pKa = 3.74AVRR243 pKa = 11.84RR244 pKa = 11.84IVEE247 pKa = 3.92PAHH250 pKa = 6.77AIVNEE255 pKa = 4.01YY256 pKa = 11.05SFDD259 pKa = 3.8PQTLSFIDD267 pKa = 3.86PAGDD271 pKa = 3.45PTYY274 pKa = 10.36HH275 pKa = 5.85ITDD278 pKa = 3.86RR279 pKa = 11.84TPSVVVAFGRR289 pKa = 11.84VLSGGSVDD297 pKa = 3.85ACSKK301 pKa = 10.97ASDD304 pKa = 4.02LRR306 pKa = 11.84TSLDD310 pKa = 3.33IPGGLPDD317 pKa = 3.28VARR320 pKa = 11.84EE321 pKa = 4.08YY322 pKa = 10.93FSSTGNVEE330 pKa = 3.82DD331 pKa = 4.88TIRR334 pKa = 11.84SAATALSDD342 pKa = 3.46FRR344 pKa = 11.84SFRR347 pKa = 11.84DD348 pKa = 3.41SHH350 pKa = 6.58PGGHH354 pKa = 6.32EE355 pKa = 4.03LNKK358 pKa = 10.82DD359 pKa = 3.15SGLSFFSRR367 pKa = 11.84DD368 pKa = 2.79RR369 pKa = 11.84ATRR372 pKa = 11.84DD373 pKa = 3.06RR374 pKa = 11.84NTAACITRR382 pKa = 11.84VEE384 pKa = 4.36EE385 pKa = 4.35VACLLRR391 pKa = 11.84SQGRR395 pKa = 11.84DD396 pKa = 2.88IARR399 pKa = 11.84MAFVVEE405 pKa = 4.03WGGPLNNDD413 pKa = 3.99SILAALALTGATACVDD429 pKa = 3.55VGPGAFKK436 pKa = 11.03VGDD439 pKa = 3.94SSEE442 pKa = 4.1EE443 pKa = 3.78LATEE447 pKa = 4.06NDD449 pKa = 3.01KK450 pKa = 11.33GAYY453 pKa = 8.7VCLIPRR459 pKa = 11.84ASTRR463 pKa = 11.84GLPRR467 pKa = 11.84MPIVGYY473 pKa = 10.41EE474 pKa = 4.13SGDD477 pKa = 3.8SVTQRR482 pKa = 11.84LDD484 pKa = 3.12RR485 pKa = 11.84LLSFISGPDD494 pKa = 3.09NYY496 pKa = 10.51PPVAYY501 pKa = 8.43ITGGSTTSGAAPADD515 pKa = 3.61IIGITQARR523 pKa = 11.84AALLTDD529 pKa = 3.66YY530 pKa = 10.42LASGSFFCGDD540 pKa = 3.0ILIPDD545 pKa = 4.6VCRR548 pKa = 11.84HH549 pKa = 6.14PPDD552 pKa = 3.94GTDD555 pKa = 4.85DD556 pKa = 3.75EE557 pKa = 5.39CLACANFDD565 pKa = 3.62YY566 pKa = 11.29SVFTVGDD573 pKa = 3.59VCQLDD578 pKa = 4.02GFSLVKK584 pKa = 10.14PRR586 pKa = 11.84ASYY589 pKa = 10.79AHH591 pKa = 6.52NLHH594 pKa = 7.1LSLEE598 pKa = 4.62WIHH601 pKa = 6.01GAARR605 pKa = 11.84EE606 pKa = 4.03FSEE609 pKa = 5.04SRR611 pKa = 11.84TLLALDD617 pKa = 3.78SVVATNVARR626 pKa = 11.84NADD629 pKa = 3.2WGDD632 pKa = 3.41FVPRR636 pKa = 11.84DD637 pKa = 3.72GPRR640 pKa = 11.84RR641 pKa = 11.84PHH643 pKa = 6.24HH644 pKa = 6.56PSYY647 pKa = 10.81GLKK650 pKa = 10.12VKK652 pKa = 10.64LIIEE656 pKa = 4.09DD657 pKa = 3.92VYY659 pKa = 11.49RR660 pKa = 11.84SMAEE664 pKa = 3.77GAEE667 pKa = 4.1EE668 pKa = 5.18GIGATKK674 pKa = 10.1TIVEE678 pKa = 4.44SLAA681 pKa = 3.57
Molecular weight: 73.28 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076JYE6|A0A076JYE6_9VIRU Putative RNA-dependent RNA polymerase OS=Cladosporium cladosporioides virus 1 OX=1529605 PE=4 SV=1
MM1 pKa = 7.68SFEE4 pKa = 4.03RR5 pKa = 11.84SAVNSWARR13 pKa = 11.84SVPVFANPDD22 pKa = 3.74SPTSPAAGRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.88EE34 pKa = 4.46SPAARR39 pKa = 11.84HH40 pKa = 5.79GATPPRR46 pKa = 11.84GRR48 pKa = 11.84PAAVNPRR55 pKa = 11.84SSGQPPPVINKK66 pKa = 9.11HH67 pKa = 4.46VTHH70 pKa = 6.41QGSSRR75 pKa = 11.84VGGVTVYY82 pKa = 10.22PGSSVSQDD90 pKa = 3.07PEE92 pKa = 3.79RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84QRR97 pKa = 11.84YY98 pKa = 7.47HH99 pKa = 6.74HH100 pKa = 6.82SPPEE104 pKa = 3.75HH105 pKa = 6.41GRR107 pKa = 11.84RR108 pKa = 11.84DD109 pKa = 3.78SSPHH113 pKa = 5.61GSGPPGSRR121 pKa = 11.84AGSSATHH128 pKa = 6.66EE129 pKa = 4.21ISRR132 pKa = 11.84GSSLHH137 pKa = 5.79RR138 pKa = 11.84RR139 pKa = 11.84ADD141 pKa = 3.62FGQRR145 pKa = 11.84AVEE148 pKa = 3.95AGRR151 pKa = 11.84PVGYY155 pKa = 9.69SSRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84DD161 pKa = 3.55TVFSDD166 pKa = 4.01AARR169 pKa = 11.84MVTVPEE175 pKa = 4.6GGFVEE180 pKa = 5.09YY181 pKa = 10.48EE182 pKa = 4.24SQTVQHH188 pKa = 6.73TDD190 pKa = 2.97GTSVTTEE197 pKa = 3.74RR198 pKa = 11.84VSVSSQGSRR207 pKa = 11.84RR208 pKa = 11.84SHH210 pKa = 6.47RR211 pKa = 11.84SNPRR215 pKa = 11.84KK216 pKa = 10.1VKK218 pKa = 9.97EE219 pKa = 4.04KK220 pKa = 10.45RR221 pKa = 11.84DD222 pKa = 3.65GNEE225 pKa = 3.45KK226 pKa = 9.82RR227 pKa = 11.84KK228 pKa = 10.48KK229 pKa = 10.5RR230 pKa = 11.84MTIGEE235 pKa = 4.18WMQTT239 pKa = 3.3
MM1 pKa = 7.68SFEE4 pKa = 4.03RR5 pKa = 11.84SAVNSWARR13 pKa = 11.84SVPVFANPDD22 pKa = 3.74SPTSPAAGRR31 pKa = 11.84RR32 pKa = 11.84EE33 pKa = 3.88EE34 pKa = 4.46SPAARR39 pKa = 11.84HH40 pKa = 5.79GATPPRR46 pKa = 11.84GRR48 pKa = 11.84PAAVNPRR55 pKa = 11.84SSGQPPPVINKK66 pKa = 9.11HH67 pKa = 4.46VTHH70 pKa = 6.41QGSSRR75 pKa = 11.84VGGVTVYY82 pKa = 10.22PGSSVSQDD90 pKa = 3.07PEE92 pKa = 3.79RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84QRR97 pKa = 11.84YY98 pKa = 7.47HH99 pKa = 6.74HH100 pKa = 6.82SPPEE104 pKa = 3.75HH105 pKa = 6.41GRR107 pKa = 11.84RR108 pKa = 11.84DD109 pKa = 3.78SSPHH113 pKa = 5.61GSGPPGSRR121 pKa = 11.84AGSSATHH128 pKa = 6.66EE129 pKa = 4.21ISRR132 pKa = 11.84GSSLHH137 pKa = 5.79RR138 pKa = 11.84RR139 pKa = 11.84ADD141 pKa = 3.62FGQRR145 pKa = 11.84AVEE148 pKa = 3.95AGRR151 pKa = 11.84PVGYY155 pKa = 9.69SSRR158 pKa = 11.84RR159 pKa = 11.84RR160 pKa = 11.84DD161 pKa = 3.55TVFSDD166 pKa = 4.01AARR169 pKa = 11.84MVTVPEE175 pKa = 4.6GGFVEE180 pKa = 5.09YY181 pKa = 10.48EE182 pKa = 4.24SQTVQHH188 pKa = 6.73TDD190 pKa = 2.97GTSVTTEE197 pKa = 3.74RR198 pKa = 11.84VSVSSQGSRR207 pKa = 11.84RR208 pKa = 11.84SHH210 pKa = 6.47RR211 pKa = 11.84SNPRR215 pKa = 11.84KK216 pKa = 10.1VKK218 pKa = 9.97EE219 pKa = 4.04KK220 pKa = 10.45RR221 pKa = 11.84DD222 pKa = 3.65GNEE225 pKa = 3.45KK226 pKa = 9.82RR227 pKa = 11.84KK228 pKa = 10.48KK229 pKa = 10.5RR230 pKa = 11.84MTIGEE235 pKa = 4.18WMQTT239 pKa = 3.3
Molecular weight: 26.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2593 |
239 |
760 |
518.6 |
55.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.567 ± 0.892 | 0.887 ± 0.235 |
6.363 ± 0.392 | 4.512 ± 0.336 |
3.625 ± 0.442 | 8.099 ± 0.379 |
2.468 ± 0.299 | 3.702 ± 0.377 |
3.355 ± 0.77 | 8.793 ± 1.101 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.315 | 2.43 ± 0.283 |
7.096 ± 0.509 | 2.198 ± 0.242 |
8.176 ± 0.702 | 9.603 ± 0.57 |
5.631 ± 0.287 | 7.443 ± 0.579 |
0.733 ± 0.132 | 2.314 ± 0.185 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
