
Synechococcus sp. (strain ATCC 27264 / PCC 7002 / PR-6) (Agmenellum quadruplicatum)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Synechococcaceae; Synechococcus; unclassified Synechococcus
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
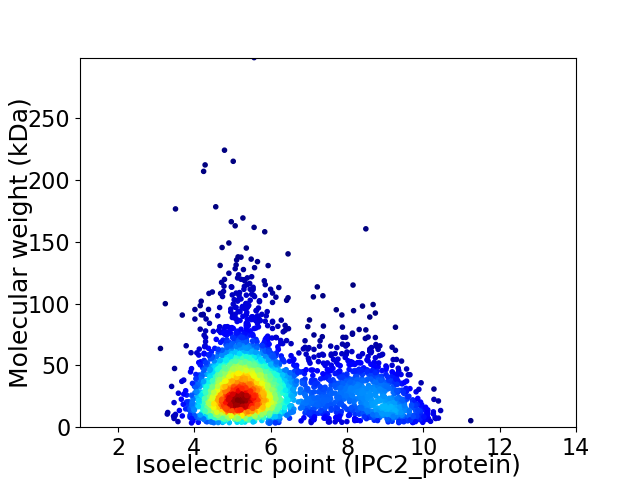
Virtual 2D-PAGE plot for 3180 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B1XJA3|B1XJA3_SYNP2 Uncharacterized protein OS=Synechococcus sp. (strain ATCC 27264 / PCC 7002 / PR-6) OX=32049 GN=SYNPCC7002_A2290 PE=4 SV=1
MM1 pKa = 7.04KK2 pKa = 10.46QSATRR7 pKa = 11.84LRR9 pKa = 11.84TLSLGLAGLTLTAALAACNTTQTPTEE35 pKa = 4.28GTGTTDD41 pKa = 3.61APADD45 pKa = 3.86SASGLSGDD53 pKa = 3.94VLIDD57 pKa = 3.61GSSTVFPITEE67 pKa = 3.69AMAEE71 pKa = 4.17EE72 pKa = 4.18FQIANPDD79 pKa = 3.13TRR81 pKa = 11.84VTVGVSGTGGGFSKK95 pKa = 10.61FCAGEE100 pKa = 3.98TDD102 pKa = 2.87ISNASRR108 pKa = 11.84PISTEE113 pKa = 3.64EE114 pKa = 4.33IEE116 pKa = 4.46ACEE119 pKa = 3.85QAGIEE124 pKa = 4.25YY125 pKa = 10.2IEE127 pKa = 4.71IPVAYY132 pKa = 9.65DD133 pKa = 4.2AISVVINPEE142 pKa = 3.97NDD144 pKa = 2.69WATCLTTEE152 pKa = 4.43EE153 pKa = 4.85LASIWSPDD161 pKa = 3.24SQINNWSQVRR171 pKa = 11.84EE172 pKa = 4.28GFPDD176 pKa = 3.89APLTLYY182 pKa = 11.07GPGTDD187 pKa = 4.59SGTFDD192 pKa = 4.07YY193 pKa = 8.83FTDD196 pKa = 5.39AINGEE201 pKa = 4.39EE202 pKa = 4.35GASRR206 pKa = 11.84GDD208 pKa = 3.59YY209 pKa = 8.26TASEE213 pKa = 4.57DD214 pKa = 4.55DD215 pKa = 3.69NVVVQGVVNDD225 pKa = 4.46PNAMGYY231 pKa = 10.26FGLAYY236 pKa = 10.57YY237 pKa = 10.05EE238 pKa = 4.39EE239 pKa = 4.61NADD242 pKa = 4.03NLNAAAIDD250 pKa = 4.91DD251 pKa = 4.55GDD253 pKa = 3.97PSNGDD258 pKa = 3.23GCVAPSTATVDD269 pKa = 3.44DD270 pKa = 4.01STYY273 pKa = 10.86QPLARR278 pKa = 11.84PIFIYY283 pKa = 10.81VSAASLDD290 pKa = 3.85EE291 pKa = 4.4KK292 pKa = 10.74PQVQAFMDD300 pKa = 4.99FYY302 pKa = 10.41TAEE305 pKa = 4.33EE306 pKa = 4.05NSSLVSEE313 pKa = 4.54VGYY316 pKa = 10.58VALNSEE322 pKa = 5.09IYY324 pKa = 10.78AKK326 pKa = 10.58AQEE329 pKa = 4.06RR330 pKa = 11.84LAARR334 pKa = 11.84TTGSIFNGGSTVGVRR349 pKa = 11.84LGEE352 pKa = 4.19VLL354 pKa = 4.05
MM1 pKa = 7.04KK2 pKa = 10.46QSATRR7 pKa = 11.84LRR9 pKa = 11.84TLSLGLAGLTLTAALAACNTTQTPTEE35 pKa = 4.28GTGTTDD41 pKa = 3.61APADD45 pKa = 3.86SASGLSGDD53 pKa = 3.94VLIDD57 pKa = 3.61GSSTVFPITEE67 pKa = 3.69AMAEE71 pKa = 4.17EE72 pKa = 4.18FQIANPDD79 pKa = 3.13TRR81 pKa = 11.84VTVGVSGTGGGFSKK95 pKa = 10.61FCAGEE100 pKa = 3.98TDD102 pKa = 2.87ISNASRR108 pKa = 11.84PISTEE113 pKa = 3.64EE114 pKa = 4.33IEE116 pKa = 4.46ACEE119 pKa = 3.85QAGIEE124 pKa = 4.25YY125 pKa = 10.2IEE127 pKa = 4.71IPVAYY132 pKa = 9.65DD133 pKa = 4.2AISVVINPEE142 pKa = 3.97NDD144 pKa = 2.69WATCLTTEE152 pKa = 4.43EE153 pKa = 4.85LASIWSPDD161 pKa = 3.24SQINNWSQVRR171 pKa = 11.84EE172 pKa = 4.28GFPDD176 pKa = 3.89APLTLYY182 pKa = 11.07GPGTDD187 pKa = 4.59SGTFDD192 pKa = 4.07YY193 pKa = 8.83FTDD196 pKa = 5.39AINGEE201 pKa = 4.39EE202 pKa = 4.35GASRR206 pKa = 11.84GDD208 pKa = 3.59YY209 pKa = 8.26TASEE213 pKa = 4.57DD214 pKa = 4.55DD215 pKa = 3.69NVVVQGVVNDD225 pKa = 4.46PNAMGYY231 pKa = 10.26FGLAYY236 pKa = 10.57YY237 pKa = 10.05EE238 pKa = 4.39EE239 pKa = 4.61NADD242 pKa = 4.03NLNAAAIDD250 pKa = 4.91DD251 pKa = 4.55GDD253 pKa = 3.97PSNGDD258 pKa = 3.23GCVAPSTATVDD269 pKa = 3.44DD270 pKa = 4.01STYY273 pKa = 10.86QPLARR278 pKa = 11.84PIFIYY283 pKa = 10.81VSAASLDD290 pKa = 3.85EE291 pKa = 4.4KK292 pKa = 10.74PQVQAFMDD300 pKa = 4.99FYY302 pKa = 10.41TAEE305 pKa = 4.33EE306 pKa = 4.05NSSLVSEE313 pKa = 4.54VGYY316 pKa = 10.58VALNSEE322 pKa = 5.09IYY324 pKa = 10.78AKK326 pKa = 10.58AQEE329 pKa = 4.06RR330 pKa = 11.84LAARR334 pKa = 11.84TTGSIFNGGSTVGVRR349 pKa = 11.84LGEE352 pKa = 4.19VLL354 pKa = 4.05
Molecular weight: 37.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|B1XJH8|RF1_SYNP2 Peptide chain release factor 1 OS=Synechococcus sp. (strain ATCC 27264 / PCC 7002 / PR-6) OX=32049 GN=prfA PE=3 SV=1
MM1 pKa = 7.55TKK3 pKa = 8.94RR4 pKa = 11.84TLGGTVRR11 pKa = 11.84KK12 pKa = 9.36QKK14 pKa = 9.22RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 7.11TGQNVIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.08KK38 pKa = 9.6GRR40 pKa = 11.84HH41 pKa = 5.24RR42 pKa = 11.84LTVV45 pKa = 3.07
MM1 pKa = 7.55TKK3 pKa = 8.94RR4 pKa = 11.84TLGGTVRR11 pKa = 11.84KK12 pKa = 9.36QKK14 pKa = 9.22RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84SHH26 pKa = 7.11TGQNVIRR33 pKa = 11.84ARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.08KK38 pKa = 9.6GRR40 pKa = 11.84HH41 pKa = 5.24RR42 pKa = 11.84LTVV45 pKa = 3.07
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
988541 |
30 |
2720 |
310.9 |
34.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.982 ± 0.047 | 1.019 ± 0.014 |
5.179 ± 0.033 | 6.14 ± 0.043 |
4.064 ± 0.034 | 7.022 ± 0.043 |
1.969 ± 0.025 | 6.283 ± 0.037 |
4.17 ± 0.038 | 11.276 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.021 | 3.739 ± 0.036 |
5.124 ± 0.034 | 5.753 ± 0.046 |
5.256 ± 0.031 | 5.474 ± 0.031 |
5.804 ± 0.035 | 6.382 ± 0.037 |
1.489 ± 0.022 | 2.936 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
