
Canis familiaris papillomavirus 17
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Taupapillomavirus; Taupapillomavirus 1
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
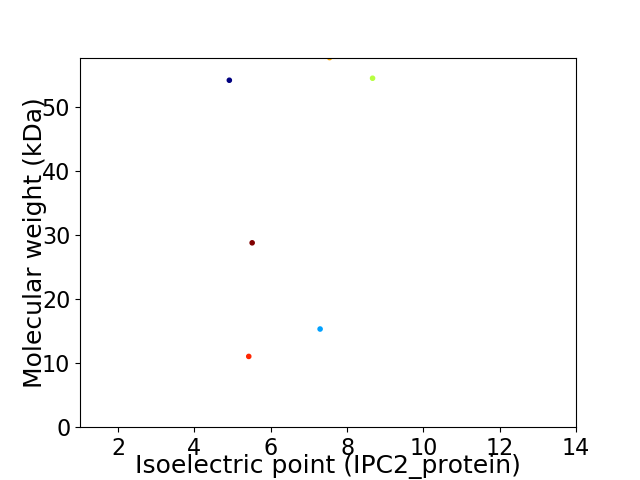
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A0U4JWN8|A0A0U4JWN8_9PAPI Regulatory protein E2 OS=Canis familiaris papillomavirus 17 OX=1778550 GN=E2 PE=3 SV=1
MM1 pKa = 7.09QPTMRR6 pKa = 11.84AKK8 pKa = 9.8RR9 pKa = 11.84VKK11 pKa = 10.08RR12 pKa = 11.84ASAEE16 pKa = 3.9QLYY19 pKa = 7.93KK20 pKa = 10.26TCQQGGDD27 pKa = 4.38CIPDD31 pKa = 3.22VVAKK35 pKa = 10.64YY36 pKa = 7.32EE37 pKa = 4.27HH38 pKa = 6.89KK39 pKa = 9.8TPADD43 pKa = 4.14KK44 pKa = 10.45ILQIGGSVIYY54 pKa = 10.68LGGLSIGTGKK64 pKa = 8.72GTGGSTGYY72 pKa = 10.33RR73 pKa = 11.84PLGVDD78 pKa = 3.33VNASTTRR85 pKa = 11.84PVLPRR90 pKa = 11.84PAIPTEE96 pKa = 4.18TIGVDD101 pKa = 3.32VGSGTVSAGEE111 pKa = 3.94SSIIPLLEE119 pKa = 4.69AGAGGDD125 pKa = 3.47AVTISPEE132 pKa = 4.0VPTITEE138 pKa = 4.17DD139 pKa = 3.98LVINPSSGAPTVTTAAGEE157 pKa = 4.53GEE159 pKa = 4.49SVAVLEE165 pKa = 4.7VGTTPSTNSRR175 pKa = 11.84VTSTSQHH182 pKa = 5.59TNPSFTPVFHH192 pKa = 6.72STPQPGEE199 pKa = 3.72ASTLEE204 pKa = 4.4SIVVGHH210 pKa = 5.21STGGRR215 pKa = 11.84VVDD218 pKa = 4.39VYY220 pKa = 11.28EE221 pKa = 5.16EE222 pKa = 4.27IPLDD226 pKa = 3.77DD227 pKa = 5.03FGPSEE232 pKa = 5.09FEE234 pKa = 4.56IEE236 pKa = 4.35DD237 pKa = 3.64PTPRR241 pKa = 11.84TSTPRR246 pKa = 11.84SAVTNLVQRR255 pKa = 11.84ARR257 pKa = 11.84QFYY260 pKa = 9.4NRR262 pKa = 11.84RR263 pKa = 11.84FAQVQVTNEE272 pKa = 3.86AFLSRR277 pKa = 11.84PASLVEE283 pKa = 4.2FEE285 pKa = 4.56NPAYY289 pKa = 10.13QPEE292 pKa = 4.35EE293 pKa = 4.16TVVFPAGSGEE303 pKa = 4.19PLAAPDD309 pKa = 4.33PDD311 pKa = 3.91FADD314 pKa = 3.24VRR316 pKa = 11.84VLNRR320 pKa = 11.84AVFQANPRR328 pKa = 11.84GGVRR332 pKa = 11.84VSRR335 pKa = 11.84LGTKK339 pKa = 8.05NTIRR343 pKa = 11.84LRR345 pKa = 11.84SGTHH349 pKa = 5.69IGGSTHH355 pKa = 5.97YY356 pKa = 10.13FYY358 pKa = 11.21DD359 pKa = 3.81LSSISLQPEE368 pKa = 3.66IEE370 pKa = 4.09LSVLGEE376 pKa = 3.99HH377 pKa = 6.41SGEE380 pKa = 4.18SEE382 pKa = 3.93IVLGSTTSTVIDD394 pKa = 3.73AVNVEE399 pKa = 4.34GTISVPEE406 pKa = 4.01EE407 pKa = 3.79DD408 pKa = 4.02VLLDD412 pKa = 3.85EE413 pKa = 4.39YY414 pKa = 11.87VEE416 pKa = 4.8DD417 pKa = 4.97FSHH420 pKa = 6.72SQLALSGGSRR430 pKa = 11.84SQSRR434 pKa = 11.84VVDD437 pKa = 3.25IPEE440 pKa = 4.06FQRR443 pKa = 11.84EE444 pKa = 4.23VPKK447 pKa = 10.93GSVIIDD453 pKa = 3.89DD454 pKa = 4.77LSDD457 pKa = 4.36SIHH460 pKa = 5.73VAHH463 pKa = 7.22PGGRR467 pKa = 11.84DD468 pKa = 3.05TTAEE472 pKa = 4.21PFSPPHH478 pKa = 6.8SSDD481 pKa = 3.68VPSSSSDD488 pKa = 3.54SVSSTFDD495 pKa = 3.1LHH497 pKa = 7.06PSLLKK502 pKa = 9.84RR503 pKa = 11.84RR504 pKa = 11.84KK505 pKa = 9.48RR506 pKa = 11.84KK507 pKa = 10.22RR508 pKa = 11.84GDD510 pKa = 2.76II511 pKa = 3.87
MM1 pKa = 7.09QPTMRR6 pKa = 11.84AKK8 pKa = 9.8RR9 pKa = 11.84VKK11 pKa = 10.08RR12 pKa = 11.84ASAEE16 pKa = 3.9QLYY19 pKa = 7.93KK20 pKa = 10.26TCQQGGDD27 pKa = 4.38CIPDD31 pKa = 3.22VVAKK35 pKa = 10.64YY36 pKa = 7.32EE37 pKa = 4.27HH38 pKa = 6.89KK39 pKa = 9.8TPADD43 pKa = 4.14KK44 pKa = 10.45ILQIGGSVIYY54 pKa = 10.68LGGLSIGTGKK64 pKa = 8.72GTGGSTGYY72 pKa = 10.33RR73 pKa = 11.84PLGVDD78 pKa = 3.33VNASTTRR85 pKa = 11.84PVLPRR90 pKa = 11.84PAIPTEE96 pKa = 4.18TIGVDD101 pKa = 3.32VGSGTVSAGEE111 pKa = 3.94SSIIPLLEE119 pKa = 4.69AGAGGDD125 pKa = 3.47AVTISPEE132 pKa = 4.0VPTITEE138 pKa = 4.17DD139 pKa = 3.98LVINPSSGAPTVTTAAGEE157 pKa = 4.53GEE159 pKa = 4.49SVAVLEE165 pKa = 4.7VGTTPSTNSRR175 pKa = 11.84VTSTSQHH182 pKa = 5.59TNPSFTPVFHH192 pKa = 6.72STPQPGEE199 pKa = 3.72ASTLEE204 pKa = 4.4SIVVGHH210 pKa = 5.21STGGRR215 pKa = 11.84VVDD218 pKa = 4.39VYY220 pKa = 11.28EE221 pKa = 5.16EE222 pKa = 4.27IPLDD226 pKa = 3.77DD227 pKa = 5.03FGPSEE232 pKa = 5.09FEE234 pKa = 4.56IEE236 pKa = 4.35DD237 pKa = 3.64PTPRR241 pKa = 11.84TSTPRR246 pKa = 11.84SAVTNLVQRR255 pKa = 11.84ARR257 pKa = 11.84QFYY260 pKa = 9.4NRR262 pKa = 11.84RR263 pKa = 11.84FAQVQVTNEE272 pKa = 3.86AFLSRR277 pKa = 11.84PASLVEE283 pKa = 4.2FEE285 pKa = 4.56NPAYY289 pKa = 10.13QPEE292 pKa = 4.35EE293 pKa = 4.16TVVFPAGSGEE303 pKa = 4.19PLAAPDD309 pKa = 4.33PDD311 pKa = 3.91FADD314 pKa = 3.24VRR316 pKa = 11.84VLNRR320 pKa = 11.84AVFQANPRR328 pKa = 11.84GGVRR332 pKa = 11.84VSRR335 pKa = 11.84LGTKK339 pKa = 8.05NTIRR343 pKa = 11.84LRR345 pKa = 11.84SGTHH349 pKa = 5.69IGGSTHH355 pKa = 5.97YY356 pKa = 10.13FYY358 pKa = 11.21DD359 pKa = 3.81LSSISLQPEE368 pKa = 3.66IEE370 pKa = 4.09LSVLGEE376 pKa = 3.99HH377 pKa = 6.41SGEE380 pKa = 4.18SEE382 pKa = 3.93IVLGSTTSTVIDD394 pKa = 3.73AVNVEE399 pKa = 4.34GTISVPEE406 pKa = 4.01EE407 pKa = 3.79DD408 pKa = 4.02VLLDD412 pKa = 3.85EE413 pKa = 4.39YY414 pKa = 11.87VEE416 pKa = 4.8DD417 pKa = 4.97FSHH420 pKa = 6.72SQLALSGGSRR430 pKa = 11.84SQSRR434 pKa = 11.84VVDD437 pKa = 3.25IPEE440 pKa = 4.06FQRR443 pKa = 11.84EE444 pKa = 4.23VPKK447 pKa = 10.93GSVIIDD453 pKa = 3.89DD454 pKa = 4.77LSDD457 pKa = 4.36SIHH460 pKa = 5.73VAHH463 pKa = 7.22PGGRR467 pKa = 11.84DD468 pKa = 3.05TTAEE472 pKa = 4.21PFSPPHH478 pKa = 6.8SSDD481 pKa = 3.68VPSSSSDD488 pKa = 3.54SVSSTFDD495 pKa = 3.1LHH497 pKa = 7.06PSLLKK502 pKa = 9.84RR503 pKa = 11.84RR504 pKa = 11.84KK505 pKa = 9.48RR506 pKa = 11.84KK507 pKa = 10.22RR508 pKa = 11.84GDD510 pKa = 2.76II511 pKa = 3.87
Molecular weight: 54.1 kDa
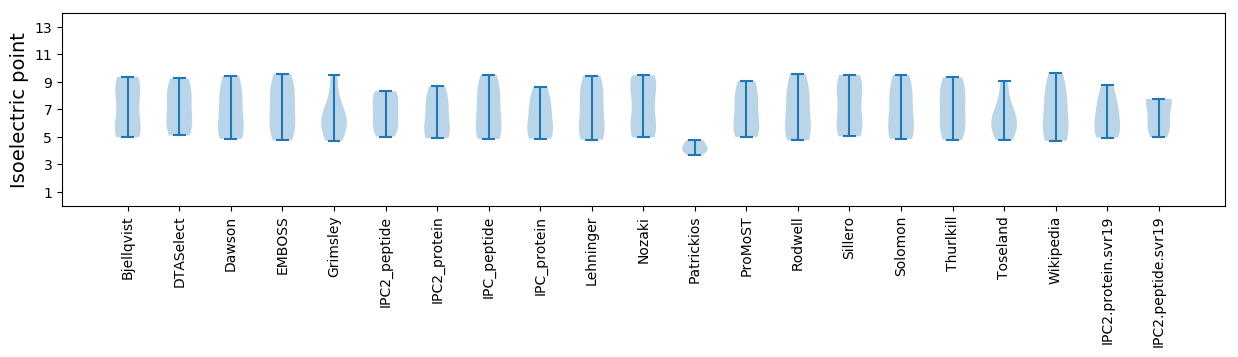
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0U4K543|A0A0U4K543_9PAPI Protein E6 OS=Canis familiaris papillomavirus 17 OX=1778550 GN=E6 PE=3 SV=1
MM1 pKa = 7.22MEE3 pKa = 4.25ALVRR7 pKa = 11.84RR8 pKa = 11.84CDD10 pKa = 3.56VLQEE14 pKa = 4.46VILQHH19 pKa = 5.74YY20 pKa = 8.31EE21 pKa = 3.91RR22 pKa = 11.84GSHH25 pKa = 6.99KK26 pKa = 10.84LSDD29 pKa = 3.29QCNYY33 pKa = 9.49WEE35 pKa = 4.02ATRR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 4.14STLLHH45 pKa = 6.43FARR48 pKa = 11.84KK49 pKa = 9.21NRR51 pKa = 11.84IHH53 pKa = 7.13RR54 pKa = 11.84LGVIPVPALATTEE67 pKa = 4.32ANAKK71 pKa = 9.9KK72 pKa = 9.93AIQMCLVLSSLNQSPYY88 pKa = 10.12RR89 pKa = 11.84DD90 pKa = 4.35EE91 pKa = 4.49PWTMTDD97 pKa = 2.9TSRR100 pKa = 11.84EE101 pKa = 3.91MWEE104 pKa = 4.45APPKK108 pKa = 10.49GCLKK112 pKa = 10.67KK113 pKa = 10.95GGVTVDD119 pKa = 3.23VLFDD123 pKa = 4.02GDD125 pKa = 4.15PNNVFPYY132 pKa = 9.29TLWSYY137 pKa = 11.14IYY139 pKa = 9.79YY140 pKa = 10.55QNLNDD145 pKa = 3.63EE146 pKa = 4.72WIKK149 pKa = 11.04VPGKK153 pKa = 10.19VDD155 pKa = 3.53YY156 pKa = 10.97EE157 pKa = 4.09GLYY160 pKa = 8.79YY161 pKa = 10.33TDD163 pKa = 4.02EE164 pKa = 5.82DD165 pKa = 4.29GEE167 pKa = 4.38KK168 pKa = 9.59IYY170 pKa = 11.08YY171 pKa = 8.94EE172 pKa = 4.18QFAKK176 pKa = 10.87DD177 pKa = 3.27AMRR180 pKa = 11.84FGNTGTWKK188 pKa = 9.91VRR190 pKa = 11.84YY191 pKa = 9.45KK192 pKa = 10.26NTVLSALVSSSGSSWPTSAPTSNSATASTTTAQRR226 pKa = 11.84EE227 pKa = 4.71GPTDD231 pKa = 3.69TPASSATSSSVSSACSSTSTGNRR254 pKa = 11.84GGPGQLIVSEE264 pKa = 4.38SQQGGSEE271 pKa = 4.17GGRR274 pKa = 11.84GRR276 pKa = 11.84GRR278 pKa = 11.84GRR280 pKa = 11.84GRR282 pKa = 11.84GRR284 pKa = 11.84GKK286 pKa = 10.56RR287 pKa = 11.84GGSPSPPLFVHH298 pKa = 6.59SDD300 pKa = 3.35WGCRR304 pKa = 11.84EE305 pKa = 3.89PQGSEE310 pKa = 3.22KK311 pKa = 10.29RR312 pKa = 11.84ARR314 pKa = 11.84EE315 pKa = 3.92ASSDD319 pKa = 3.78GEE321 pKa = 5.34DD322 pKa = 3.15RR323 pKa = 11.84ATWGGGRR330 pKa = 11.84EE331 pKa = 3.7RR332 pKa = 11.84RR333 pKa = 11.84AGQGEE338 pKa = 4.17RR339 pKa = 11.84PSCSSCNLSPNPSTIPTLSTPGPTPGHH366 pKa = 5.33GHH368 pKa = 7.03GGRR371 pKa = 11.84PTPKK375 pKa = 10.26APRR378 pKa = 11.84LSPAVPLEE386 pKa = 3.88QLVQRR391 pKa = 11.84PGDD394 pKa = 3.5FEE396 pKa = 4.73TEE398 pKa = 3.72VGRR401 pKa = 11.84RR402 pKa = 11.84PRR404 pKa = 11.84GPEE407 pKa = 3.81GQNRR411 pKa = 11.84PYY413 pKa = 10.47PVIIVKK419 pKa = 10.7GDD421 pKa = 3.42GNASKK426 pKa = 10.44CWRR429 pKa = 11.84RR430 pKa = 11.84RR431 pKa = 11.84LRR433 pKa = 11.84LRR435 pKa = 11.84YY436 pKa = 9.82GEE438 pKa = 5.19LYY440 pKa = 9.78TAISTGFSWTTANGPHH456 pKa = 6.22RR457 pKa = 11.84VCRR460 pKa = 11.84PRR462 pKa = 11.84MVIAFDD468 pKa = 3.93SDD470 pKa = 3.4EE471 pKa = 3.85QRR473 pKa = 11.84AAFLKK478 pKa = 10.31NVRR481 pKa = 11.84IPKK484 pKa = 9.95VLDD487 pKa = 3.1IAFGSLDD494 pKa = 3.52SLL496 pKa = 4.34
MM1 pKa = 7.22MEE3 pKa = 4.25ALVRR7 pKa = 11.84RR8 pKa = 11.84CDD10 pKa = 3.56VLQEE14 pKa = 4.46VILQHH19 pKa = 5.74YY20 pKa = 8.31EE21 pKa = 3.91RR22 pKa = 11.84GSHH25 pKa = 6.99KK26 pKa = 10.84LSDD29 pKa = 3.29QCNYY33 pKa = 9.49WEE35 pKa = 4.02ATRR38 pKa = 11.84RR39 pKa = 11.84EE40 pKa = 4.14STLLHH45 pKa = 6.43FARR48 pKa = 11.84KK49 pKa = 9.21NRR51 pKa = 11.84IHH53 pKa = 7.13RR54 pKa = 11.84LGVIPVPALATTEE67 pKa = 4.32ANAKK71 pKa = 9.9KK72 pKa = 9.93AIQMCLVLSSLNQSPYY88 pKa = 10.12RR89 pKa = 11.84DD90 pKa = 4.35EE91 pKa = 4.49PWTMTDD97 pKa = 2.9TSRR100 pKa = 11.84EE101 pKa = 3.91MWEE104 pKa = 4.45APPKK108 pKa = 10.49GCLKK112 pKa = 10.67KK113 pKa = 10.95GGVTVDD119 pKa = 3.23VLFDD123 pKa = 4.02GDD125 pKa = 4.15PNNVFPYY132 pKa = 9.29TLWSYY137 pKa = 11.14IYY139 pKa = 9.79YY140 pKa = 10.55QNLNDD145 pKa = 3.63EE146 pKa = 4.72WIKK149 pKa = 11.04VPGKK153 pKa = 10.19VDD155 pKa = 3.53YY156 pKa = 10.97EE157 pKa = 4.09GLYY160 pKa = 8.79YY161 pKa = 10.33TDD163 pKa = 4.02EE164 pKa = 5.82DD165 pKa = 4.29GEE167 pKa = 4.38KK168 pKa = 9.59IYY170 pKa = 11.08YY171 pKa = 8.94EE172 pKa = 4.18QFAKK176 pKa = 10.87DD177 pKa = 3.27AMRR180 pKa = 11.84FGNTGTWKK188 pKa = 9.91VRR190 pKa = 11.84YY191 pKa = 9.45KK192 pKa = 10.26NTVLSALVSSSGSSWPTSAPTSNSATASTTTAQRR226 pKa = 11.84EE227 pKa = 4.71GPTDD231 pKa = 3.69TPASSATSSSVSSACSSTSTGNRR254 pKa = 11.84GGPGQLIVSEE264 pKa = 4.38SQQGGSEE271 pKa = 4.17GGRR274 pKa = 11.84GRR276 pKa = 11.84GRR278 pKa = 11.84GRR280 pKa = 11.84GRR282 pKa = 11.84GRR284 pKa = 11.84GKK286 pKa = 10.56RR287 pKa = 11.84GGSPSPPLFVHH298 pKa = 6.59SDD300 pKa = 3.35WGCRR304 pKa = 11.84EE305 pKa = 3.89PQGSEE310 pKa = 3.22KK311 pKa = 10.29RR312 pKa = 11.84ARR314 pKa = 11.84EE315 pKa = 3.92ASSDD319 pKa = 3.78GEE321 pKa = 5.34DD322 pKa = 3.15RR323 pKa = 11.84ATWGGGRR330 pKa = 11.84EE331 pKa = 3.7RR332 pKa = 11.84RR333 pKa = 11.84AGQGEE338 pKa = 4.17RR339 pKa = 11.84PSCSSCNLSPNPSTIPTLSTPGPTPGHH366 pKa = 5.33GHH368 pKa = 7.03GGRR371 pKa = 11.84PTPKK375 pKa = 10.26APRR378 pKa = 11.84LSPAVPLEE386 pKa = 3.88QLVQRR391 pKa = 11.84PGDD394 pKa = 3.5FEE396 pKa = 4.73TEE398 pKa = 3.72VGRR401 pKa = 11.84RR402 pKa = 11.84PRR404 pKa = 11.84GPEE407 pKa = 3.81GQNRR411 pKa = 11.84PYY413 pKa = 10.47PVIIVKK419 pKa = 10.7GDD421 pKa = 3.42GNASKK426 pKa = 10.44CWRR429 pKa = 11.84RR430 pKa = 11.84RR431 pKa = 11.84LRR433 pKa = 11.84LRR435 pKa = 11.84YY436 pKa = 9.82GEE438 pKa = 5.19LYY440 pKa = 9.78TAISTGFSWTTANGPHH456 pKa = 6.22RR457 pKa = 11.84VCRR460 pKa = 11.84PRR462 pKa = 11.84MVIAFDD468 pKa = 3.93SDD470 pKa = 3.4EE471 pKa = 3.85QRR473 pKa = 11.84AAFLKK478 pKa = 10.31NVRR481 pKa = 11.84IPKK484 pKa = 9.95VLDD487 pKa = 3.1IAFGSLDD494 pKa = 3.52SLL496 pKa = 4.34
Molecular weight: 54.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2006 |
98 |
511 |
334.3 |
36.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.334 ± 0.479 | 2.293 ± 0.79 |
5.733 ± 0.487 | 6.431 ± 0.568 |
3.24 ± 0.453 | 8.475 ± 0.737 |
2.044 ± 0.359 | 4.038 ± 0.409 |
4.487 ± 0.922 | 7.527 ± 0.574 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.496 ± 0.407 | 3.639 ± 0.741 |
8.624 ± 1.438 | 3.789 ± 0.434 |
7.228 ± 0.834 | 8.225 ± 1.338 |
6.78 ± 0.787 | 6.73 ± 1.234 |
1.196 ± 0.394 | 2.692 ± 0.626 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
