
Ornithinimicrobium cerasi
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Ornithinimicrobiaceae; Ornithinimicrobium
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
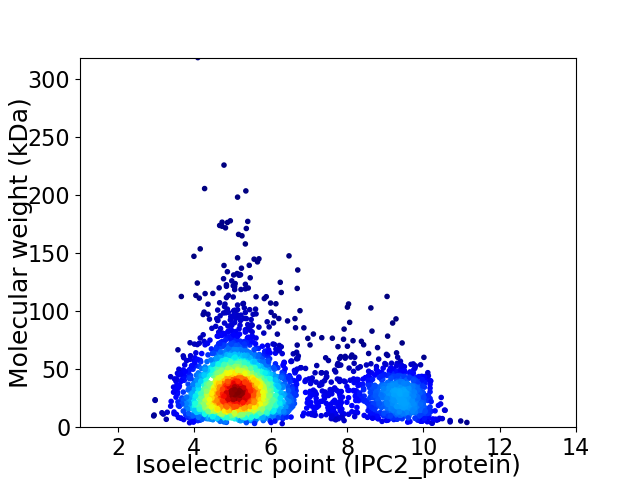
Virtual 2D-PAGE plot for 3368 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
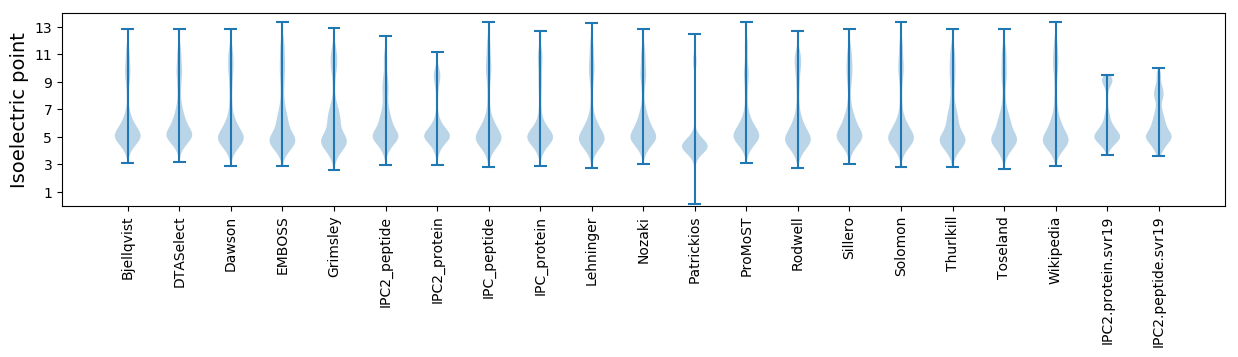
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285VPA4|A0A285VPA4_9MICO Proteasome accessory factor C OS=Ornithinimicrobium cerasi OX=2248773 GN=SAMN05421879_103127 PE=4 SV=1
MM1 pKa = 7.96RR2 pKa = 11.84NRR4 pKa = 11.84SVGALASAAALALLLTSCAEE24 pKa = 4.15SDD26 pKa = 4.41RR27 pKa = 11.84DD28 pKa = 3.85TGDD31 pKa = 4.43DD32 pKa = 3.95GDD34 pKa = 4.32GAPAGTDD41 pKa = 2.84NATGTEE47 pKa = 4.09PAAGGDD53 pKa = 4.22DD54 pKa = 3.84GASDD58 pKa = 3.47ATFVFGAAGAPTTFDD73 pKa = 3.41PFYY76 pKa = 10.83ASDD79 pKa = 3.89GEE81 pKa = 4.57TFRR84 pKa = 11.84VTRR87 pKa = 11.84QIFQNLIGIEE97 pKa = 4.15EE98 pKa = 4.7GGSEE102 pKa = 4.56AVPEE106 pKa = 4.6LATSWEE112 pKa = 4.35SEE114 pKa = 4.48DD115 pKa = 3.63GLVWDD120 pKa = 5.03FAIQEE125 pKa = 4.44GVTFHH130 pKa = 7.79DD131 pKa = 4.43GTPLDD136 pKa = 4.36AEE138 pKa = 4.32AVCANFTRR146 pKa = 11.84WADD149 pKa = 3.53QNEE152 pKa = 4.38AGQSPSGAYY161 pKa = 10.26YY162 pKa = 10.1YY163 pKa = 11.4GNDD166 pKa = 3.25FGFGEE171 pKa = 4.41DD172 pKa = 4.31SLYY175 pKa = 10.66EE176 pKa = 4.13SCEE179 pKa = 3.95ATDD182 pKa = 3.74EE183 pKa = 4.1LTAQVTVSRR192 pKa = 11.84VTGKK196 pKa = 10.49FPMVLSQSSYY206 pKa = 11.1AIQSPTAMDD215 pKa = 4.01EE216 pKa = 3.9YY217 pKa = 10.69DD218 pKa = 3.55ANGIALEE225 pKa = 4.46GEE227 pKa = 3.91AFVFPEE233 pKa = 4.11YY234 pKa = 10.63AQEE237 pKa = 4.44HH238 pKa = 5.6PTGTGPFMFEE248 pKa = 4.39SYY250 pKa = 11.19DD251 pKa = 3.6NAGGSVTLTRR261 pKa = 11.84NDD263 pKa = 4.79DD264 pKa = 3.57YY265 pKa = 11.2WGDD268 pKa = 3.51PAGVSEE274 pKa = 4.28IVFRR278 pKa = 11.84IIPDD282 pKa = 3.23EE283 pKa = 3.75NARR286 pKa = 11.84RR287 pKa = 11.84QEE289 pKa = 4.13LEE291 pKa = 3.65AGTINGYY298 pKa = 9.2DD299 pKa = 3.71LPNPVDD305 pKa = 3.37WQALEE310 pKa = 4.38DD311 pKa = 4.17AGNQVLIRR319 pKa = 11.84DD320 pKa = 4.45PFNILYY326 pKa = 10.26LALNPVADD334 pKa = 4.48PQLEE338 pKa = 4.38DD339 pKa = 3.62PLVRR343 pKa = 11.84QALYY347 pKa = 10.27HH348 pKa = 6.02ALNRR352 pKa = 11.84QQFVTSQLPAGAEE365 pKa = 4.06VATQFMPSTVSGWSSGIDD383 pKa = 2.99AYY385 pKa = 10.52EE386 pKa = 3.93YY387 pKa = 10.7DD388 pKa = 3.74VEE390 pKa = 4.26KK391 pKa = 11.08ARR393 pKa = 11.84DD394 pKa = 3.67LLEE397 pKa = 4.04QAGKK401 pKa = 10.5SDD403 pKa = 3.6LTIEE407 pKa = 4.21LWYY410 pKa = 9.4PSEE413 pKa = 4.0VTRR416 pKa = 11.84PYY418 pKa = 10.19MPDD421 pKa = 2.98PTRR424 pKa = 11.84VYY426 pKa = 10.98DD427 pKa = 3.68AVKK430 pKa = 10.61ADD432 pKa = 3.51WEE434 pKa = 4.27AAGITVEE441 pKa = 4.5TVTKK445 pKa = 8.82PWAGGYY451 pKa = 9.64IDD453 pKa = 4.97DD454 pKa = 4.6TQASRR459 pKa = 11.84APAFFLGWTGDD470 pKa = 3.82LNSADD475 pKa = 3.93NFLCAFFCGDD485 pKa = 3.94DD486 pKa = 3.78NQFGTSAYY494 pKa = 9.85DD495 pKa = 3.08WHH497 pKa = 7.45EE498 pKa = 4.35DD499 pKa = 3.48LQEE502 pKa = 4.08QIAAADD508 pKa = 3.82AEE510 pKa = 4.4ADD512 pKa = 3.49EE513 pKa = 4.46ATRR516 pKa = 11.84TGLYY520 pKa = 10.06EE521 pKa = 4.27SLNEE525 pKa = 4.68AIMSPEE531 pKa = 4.11WLPGLPISHH540 pKa = 6.89SPPAIVVGEE549 pKa = 4.08NVQGLVASPLTAEE562 pKa = 4.84DD563 pKa = 4.57FSTVTITEE571 pKa = 3.98
MM1 pKa = 7.96RR2 pKa = 11.84NRR4 pKa = 11.84SVGALASAAALALLLTSCAEE24 pKa = 4.15SDD26 pKa = 4.41RR27 pKa = 11.84DD28 pKa = 3.85TGDD31 pKa = 4.43DD32 pKa = 3.95GDD34 pKa = 4.32GAPAGTDD41 pKa = 2.84NATGTEE47 pKa = 4.09PAAGGDD53 pKa = 4.22DD54 pKa = 3.84GASDD58 pKa = 3.47ATFVFGAAGAPTTFDD73 pKa = 3.41PFYY76 pKa = 10.83ASDD79 pKa = 3.89GEE81 pKa = 4.57TFRR84 pKa = 11.84VTRR87 pKa = 11.84QIFQNLIGIEE97 pKa = 4.15EE98 pKa = 4.7GGSEE102 pKa = 4.56AVPEE106 pKa = 4.6LATSWEE112 pKa = 4.35SEE114 pKa = 4.48DD115 pKa = 3.63GLVWDD120 pKa = 5.03FAIQEE125 pKa = 4.44GVTFHH130 pKa = 7.79DD131 pKa = 4.43GTPLDD136 pKa = 4.36AEE138 pKa = 4.32AVCANFTRR146 pKa = 11.84WADD149 pKa = 3.53QNEE152 pKa = 4.38AGQSPSGAYY161 pKa = 10.26YY162 pKa = 10.1YY163 pKa = 11.4GNDD166 pKa = 3.25FGFGEE171 pKa = 4.41DD172 pKa = 4.31SLYY175 pKa = 10.66EE176 pKa = 4.13SCEE179 pKa = 3.95ATDD182 pKa = 3.74EE183 pKa = 4.1LTAQVTVSRR192 pKa = 11.84VTGKK196 pKa = 10.49FPMVLSQSSYY206 pKa = 11.1AIQSPTAMDD215 pKa = 4.01EE216 pKa = 3.9YY217 pKa = 10.69DD218 pKa = 3.55ANGIALEE225 pKa = 4.46GEE227 pKa = 3.91AFVFPEE233 pKa = 4.11YY234 pKa = 10.63AQEE237 pKa = 4.44HH238 pKa = 5.6PTGTGPFMFEE248 pKa = 4.39SYY250 pKa = 11.19DD251 pKa = 3.6NAGGSVTLTRR261 pKa = 11.84NDD263 pKa = 4.79DD264 pKa = 3.57YY265 pKa = 11.2WGDD268 pKa = 3.51PAGVSEE274 pKa = 4.28IVFRR278 pKa = 11.84IIPDD282 pKa = 3.23EE283 pKa = 3.75NARR286 pKa = 11.84RR287 pKa = 11.84QEE289 pKa = 4.13LEE291 pKa = 3.65AGTINGYY298 pKa = 9.2DD299 pKa = 3.71LPNPVDD305 pKa = 3.37WQALEE310 pKa = 4.38DD311 pKa = 4.17AGNQVLIRR319 pKa = 11.84DD320 pKa = 4.45PFNILYY326 pKa = 10.26LALNPVADD334 pKa = 4.48PQLEE338 pKa = 4.38DD339 pKa = 3.62PLVRR343 pKa = 11.84QALYY347 pKa = 10.27HH348 pKa = 6.02ALNRR352 pKa = 11.84QQFVTSQLPAGAEE365 pKa = 4.06VATQFMPSTVSGWSSGIDD383 pKa = 2.99AYY385 pKa = 10.52EE386 pKa = 3.93YY387 pKa = 10.7DD388 pKa = 3.74VEE390 pKa = 4.26KK391 pKa = 11.08ARR393 pKa = 11.84DD394 pKa = 3.67LLEE397 pKa = 4.04QAGKK401 pKa = 10.5SDD403 pKa = 3.6LTIEE407 pKa = 4.21LWYY410 pKa = 9.4PSEE413 pKa = 4.0VTRR416 pKa = 11.84PYY418 pKa = 10.19MPDD421 pKa = 2.98PTRR424 pKa = 11.84VYY426 pKa = 10.98DD427 pKa = 3.68AVKK430 pKa = 10.61ADD432 pKa = 3.51WEE434 pKa = 4.27AAGITVEE441 pKa = 4.5TVTKK445 pKa = 8.82PWAGGYY451 pKa = 9.64IDD453 pKa = 4.97DD454 pKa = 4.6TQASRR459 pKa = 11.84APAFFLGWTGDD470 pKa = 3.82LNSADD475 pKa = 3.93NFLCAFFCGDD485 pKa = 3.94DD486 pKa = 3.78NQFGTSAYY494 pKa = 9.85DD495 pKa = 3.08WHH497 pKa = 7.45EE498 pKa = 4.35DD499 pKa = 3.48LQEE502 pKa = 4.08QIAAADD508 pKa = 3.82AEE510 pKa = 4.4ADD512 pKa = 3.49EE513 pKa = 4.46ATRR516 pKa = 11.84TGLYY520 pKa = 10.06EE521 pKa = 4.27SLNEE525 pKa = 4.68AIMSPEE531 pKa = 4.11WLPGLPISHH540 pKa = 6.89SPPAIVVGEE549 pKa = 4.08NVQGLVASPLTAEE562 pKa = 4.84DD563 pKa = 4.57FSTVTITEE571 pKa = 3.98
Molecular weight: 61.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285VUX1|A0A285VUX1_9MICO Lysophospholipase L1 OS=Ornithinimicrobium cerasi OX=2248773 GN=SAMN05421879_11721 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1132626 |
27 |
2986 |
336.3 |
35.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.954 ± 0.057 | 0.638 ± 0.011 |
6.374 ± 0.038 | 6.034 ± 0.039 |
2.444 ± 0.025 | 9.65 ± 0.042 |
2.291 ± 0.02 | 2.78 ± 0.03 |
1.356 ± 0.026 | 10.672 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.805 ± 0.016 | 1.45 ± 0.024 |
5.961 ± 0.036 | 2.868 ± 0.023 |
8.157 ± 0.046 | 5.015 ± 0.026 |
6.135 ± 0.034 | 9.947 ± 0.047 |
1.64 ± 0.017 | 1.828 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
