
Humitalea rosea
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Acetobacteraceae; Humitalea
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
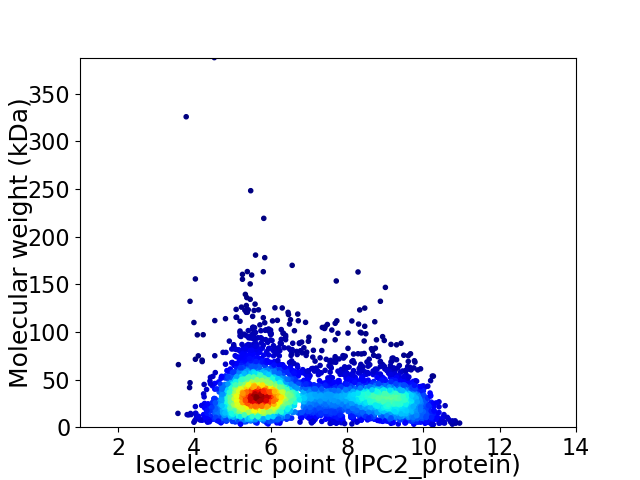
Virtual 2D-PAGE plot for 4650 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
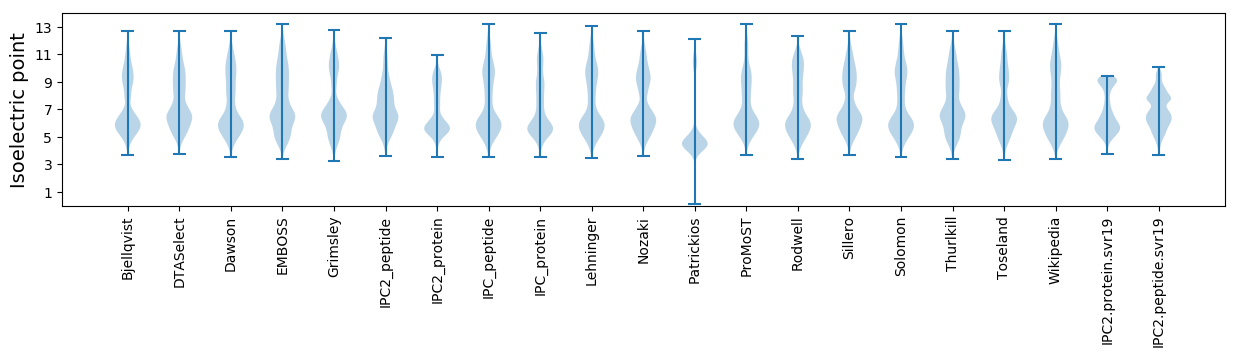
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W7IT37|A0A2W7IT37_9PROT Cell shape-determining protein MreB OS=Humitalea rosea OX=990373 GN=mreB PE=3 SV=1
MM1 pKa = 7.63ARR3 pKa = 11.84FTLTEE8 pKa = 3.89NALITPQVPGFARR21 pKa = 11.84LDD23 pKa = 3.63LLATITLPNVSDD35 pKa = 3.87AYY37 pKa = 10.96VYY39 pKa = 11.35GDD41 pKa = 4.01FRR43 pKa = 11.84ASGQAAYY50 pKa = 10.22SVEE53 pKa = 3.79FLGFGLTAANGRR65 pKa = 11.84LSDD68 pKa = 3.84GTITALKK75 pKa = 8.32YY76 pKa = 8.57TLSGIQVFYY85 pKa = 11.28LDD87 pKa = 5.13LLQLPVATFNAYY99 pKa = 9.17IDD101 pKa = 3.99SRR103 pKa = 11.84DD104 pKa = 3.44EE105 pKa = 4.46AGFATYY111 pKa = 10.53LFSGNDD117 pKa = 3.33EE118 pKa = 4.6LYY120 pKa = 8.49GTSRR124 pKa = 11.84ADD126 pKa = 3.42LLTGYY131 pKa = 10.54GGDD134 pKa = 3.99DD135 pKa = 3.72VIGGKK140 pKa = 9.98DD141 pKa = 3.74GNDD144 pKa = 3.18TLAGGEE150 pKa = 4.51GNDD153 pKa = 3.78TLYY156 pKa = 11.48GEE158 pKa = 4.98ADD160 pKa = 3.3NDD162 pKa = 3.66VLYY165 pKa = 10.81GAGGADD171 pKa = 3.52EE172 pKa = 4.45VHH174 pKa = 6.66GGTGNDD180 pKa = 4.38FIYY183 pKa = 10.61LGEE186 pKa = 4.31GADD189 pKa = 3.58NGFGEE194 pKa = 5.01AGNDD198 pKa = 3.74TIFGEE203 pKa = 4.54GGNDD207 pKa = 3.32IIAGGVEE214 pKa = 3.77NDD216 pKa = 4.36YY217 pKa = 11.32IDD219 pKa = 5.16GGNGDD224 pKa = 5.35DD225 pKa = 4.31NLQGEE230 pKa = 4.86AGNDD234 pKa = 3.69TLRR237 pKa = 11.84GGAGNDD243 pKa = 3.83SISGGVEE250 pKa = 3.43EE251 pKa = 5.07DD252 pKa = 4.04LVYY255 pKa = 11.23GDD257 pKa = 5.14AGNDD261 pKa = 3.64SIYY264 pKa = 10.74LDD266 pKa = 3.91SGQDD270 pKa = 3.15TAYY273 pKa = 10.64GGTGDD278 pKa = 4.0DD279 pKa = 4.53TIFGDD284 pKa = 3.54TGDD287 pKa = 4.23DD288 pKa = 3.65VLVGEE293 pKa = 5.59DD294 pKa = 4.73GDD296 pKa = 4.14DD297 pKa = 3.59QLFGGNGNDD306 pKa = 4.16TILGGAGADD315 pKa = 3.22QLFGGEE321 pKa = 4.72NLDD324 pKa = 4.83LLYY327 pKa = 11.24GDD329 pKa = 4.73IGNDD333 pKa = 3.18TLYY336 pKa = 11.39GEE338 pKa = 5.14GSFDD342 pKa = 3.5TAGVSTLWRR351 pKa = 11.84QNTLTGTVGNFTLTGPEE368 pKa = 4.27GVDD371 pKa = 2.94TLYY374 pKa = 10.91SIEE377 pKa = 4.75SIRR380 pKa = 11.84SLDD383 pKa = 3.08GWLEE387 pKa = 3.91LDD389 pKa = 3.28GGGPVSQVARR399 pKa = 11.84LYY401 pKa = 10.92LATLGRR407 pKa = 11.84ASDD410 pKa = 3.79ATGLGYY416 pKa = 10.57YY417 pKa = 9.4VAAIEE422 pKa = 5.67AGTQTLNDD430 pKa = 3.26ISAGLVASAEE440 pKa = 3.96FGARR444 pKa = 11.84YY445 pKa = 9.47GNPGNSDD452 pKa = 3.85FVTLLYY458 pKa = 11.19ANVLNRR464 pKa = 11.84APDD467 pKa = 3.6SGGLAYY473 pKa = 10.43YY474 pKa = 9.43VARR477 pKa = 11.84LDD479 pKa = 4.07AGEE482 pKa = 3.94QRR484 pKa = 11.84STIVLDD490 pKa = 4.38FSEE493 pKa = 4.71SDD495 pKa = 3.29EE496 pKa = 4.6FIYY499 pKa = 10.61RR500 pKa = 11.84AGYY503 pKa = 8.66PGEE506 pKa = 4.36GGIFAPDD513 pKa = 3.68PVAVDD518 pKa = 3.2VVRR521 pKa = 11.84YY522 pKa = 9.64YY523 pKa = 10.24EE524 pKa = 4.27TVLDD528 pKa = 4.29RR529 pKa = 11.84LPDD532 pKa = 4.01AGGAAYY538 pKa = 8.75WIQLRR543 pKa = 11.84HH544 pKa = 6.08GGLSLANMGAAFTGSAEE561 pKa = 3.92FEE563 pKa = 4.22SRR565 pKa = 11.84YY566 pKa = 10.16GALSNSGFVEE576 pKa = 3.83QLYY579 pKa = 11.23LNALDD584 pKa = 4.13RR585 pKa = 11.84AGDD588 pKa = 3.77SGGIAYY594 pKa = 7.21WTAEE598 pKa = 4.24LDD600 pKa = 3.27HH601 pKa = 6.68GALSRR606 pKa = 11.84AGVADD611 pKa = 3.69SFAFSAEE618 pKa = 3.8MTAKK622 pKa = 10.39VEE624 pKa = 3.99PLVDD628 pKa = 3.68GGITFAA634 pKa = 5.51
MM1 pKa = 7.63ARR3 pKa = 11.84FTLTEE8 pKa = 3.89NALITPQVPGFARR21 pKa = 11.84LDD23 pKa = 3.63LLATITLPNVSDD35 pKa = 3.87AYY37 pKa = 10.96VYY39 pKa = 11.35GDD41 pKa = 4.01FRR43 pKa = 11.84ASGQAAYY50 pKa = 10.22SVEE53 pKa = 3.79FLGFGLTAANGRR65 pKa = 11.84LSDD68 pKa = 3.84GTITALKK75 pKa = 8.32YY76 pKa = 8.57TLSGIQVFYY85 pKa = 11.28LDD87 pKa = 5.13LLQLPVATFNAYY99 pKa = 9.17IDD101 pKa = 3.99SRR103 pKa = 11.84DD104 pKa = 3.44EE105 pKa = 4.46AGFATYY111 pKa = 10.53LFSGNDD117 pKa = 3.33EE118 pKa = 4.6LYY120 pKa = 8.49GTSRR124 pKa = 11.84ADD126 pKa = 3.42LLTGYY131 pKa = 10.54GGDD134 pKa = 3.99DD135 pKa = 3.72VIGGKK140 pKa = 9.98DD141 pKa = 3.74GNDD144 pKa = 3.18TLAGGEE150 pKa = 4.51GNDD153 pKa = 3.78TLYY156 pKa = 11.48GEE158 pKa = 4.98ADD160 pKa = 3.3NDD162 pKa = 3.66VLYY165 pKa = 10.81GAGGADD171 pKa = 3.52EE172 pKa = 4.45VHH174 pKa = 6.66GGTGNDD180 pKa = 4.38FIYY183 pKa = 10.61LGEE186 pKa = 4.31GADD189 pKa = 3.58NGFGEE194 pKa = 5.01AGNDD198 pKa = 3.74TIFGEE203 pKa = 4.54GGNDD207 pKa = 3.32IIAGGVEE214 pKa = 3.77NDD216 pKa = 4.36YY217 pKa = 11.32IDD219 pKa = 5.16GGNGDD224 pKa = 5.35DD225 pKa = 4.31NLQGEE230 pKa = 4.86AGNDD234 pKa = 3.69TLRR237 pKa = 11.84GGAGNDD243 pKa = 3.83SISGGVEE250 pKa = 3.43EE251 pKa = 5.07DD252 pKa = 4.04LVYY255 pKa = 11.23GDD257 pKa = 5.14AGNDD261 pKa = 3.64SIYY264 pKa = 10.74LDD266 pKa = 3.91SGQDD270 pKa = 3.15TAYY273 pKa = 10.64GGTGDD278 pKa = 4.0DD279 pKa = 4.53TIFGDD284 pKa = 3.54TGDD287 pKa = 4.23DD288 pKa = 3.65VLVGEE293 pKa = 5.59DD294 pKa = 4.73GDD296 pKa = 4.14DD297 pKa = 3.59QLFGGNGNDD306 pKa = 4.16TILGGAGADD315 pKa = 3.22QLFGGEE321 pKa = 4.72NLDD324 pKa = 4.83LLYY327 pKa = 11.24GDD329 pKa = 4.73IGNDD333 pKa = 3.18TLYY336 pKa = 11.39GEE338 pKa = 5.14GSFDD342 pKa = 3.5TAGVSTLWRR351 pKa = 11.84QNTLTGTVGNFTLTGPEE368 pKa = 4.27GVDD371 pKa = 2.94TLYY374 pKa = 10.91SIEE377 pKa = 4.75SIRR380 pKa = 11.84SLDD383 pKa = 3.08GWLEE387 pKa = 3.91LDD389 pKa = 3.28GGGPVSQVARR399 pKa = 11.84LYY401 pKa = 10.92LATLGRR407 pKa = 11.84ASDD410 pKa = 3.79ATGLGYY416 pKa = 10.57YY417 pKa = 9.4VAAIEE422 pKa = 5.67AGTQTLNDD430 pKa = 3.26ISAGLVASAEE440 pKa = 3.96FGARR444 pKa = 11.84YY445 pKa = 9.47GNPGNSDD452 pKa = 3.85FVTLLYY458 pKa = 11.19ANVLNRR464 pKa = 11.84APDD467 pKa = 3.6SGGLAYY473 pKa = 10.43YY474 pKa = 9.43VARR477 pKa = 11.84LDD479 pKa = 4.07AGEE482 pKa = 3.94QRR484 pKa = 11.84STIVLDD490 pKa = 4.38FSEE493 pKa = 4.71SDD495 pKa = 3.29EE496 pKa = 4.6FIYY499 pKa = 10.61RR500 pKa = 11.84AGYY503 pKa = 8.66PGEE506 pKa = 4.36GGIFAPDD513 pKa = 3.68PVAVDD518 pKa = 3.2VVRR521 pKa = 11.84YY522 pKa = 9.64YY523 pKa = 10.24EE524 pKa = 4.27TVLDD528 pKa = 4.29RR529 pKa = 11.84LPDD532 pKa = 4.01AGGAAYY538 pKa = 8.75WIQLRR543 pKa = 11.84HH544 pKa = 6.08GGLSLANMGAAFTGSAEE561 pKa = 3.92FEE563 pKa = 4.22SRR565 pKa = 11.84YY566 pKa = 10.16GALSNSGFVEE576 pKa = 3.83QLYY579 pKa = 11.23LNALDD584 pKa = 4.13RR585 pKa = 11.84AGDD588 pKa = 3.77SGGIAYY594 pKa = 7.21WTAEE598 pKa = 4.24LDD600 pKa = 3.27HH601 pKa = 6.68GALSRR606 pKa = 11.84AGVADD611 pKa = 3.69SFAFSAEE618 pKa = 3.8MTAKK622 pKa = 10.39VEE624 pKa = 3.99PLVDD628 pKa = 3.68GGITFAA634 pKa = 5.51
Molecular weight: 65.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W7I6H0|A0A2W7I6H0_9PROT Formate dehydrogenase gamma subunit OS=Humitalea rosea OX=990373 GN=C8P66_11840 PE=3 SV=1
GG1 pKa = 7.01KK2 pKa = 9.51RR3 pKa = 11.84HH4 pKa = 5.45HH5 pKa = 6.27QAVIALARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VNVLWAVVQNRR26 pKa = 11.84TPFQTRR32 pKa = 11.84FKK34 pKa = 10.09MAAA37 pKa = 3.11
GG1 pKa = 7.01KK2 pKa = 9.51RR3 pKa = 11.84HH4 pKa = 5.45HH5 pKa = 6.27QAVIALARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VNVLWAVVQNRR26 pKa = 11.84TPFQTRR32 pKa = 11.84FKK34 pKa = 10.09MAAA37 pKa = 3.11
Molecular weight: 4.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1537676 |
25 |
4046 |
330.7 |
35.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.091 ± 0.064 | 0.853 ± 0.011 |
5.115 ± 0.024 | 5.145 ± 0.03 |
3.311 ± 0.02 | 9.335 ± 0.039 |
1.976 ± 0.016 | 4.384 ± 0.021 |
1.864 ± 0.023 | 11.071 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.476 ± 0.017 | 1.991 ± 0.016 |
6.255 ± 0.032 | 2.867 ± 0.02 |
7.986 ± 0.033 | 4.502 ± 0.022 |
5.178 ± 0.034 | 7.417 ± 0.03 |
1.453 ± 0.015 | 1.73 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
