
Pseudomassariella vexata
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; sordariomyceta; Sordariomycetes; Xylariomycetidae; Xylariales; Pseudomassariaceae; Pseudomassariella
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 12557 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
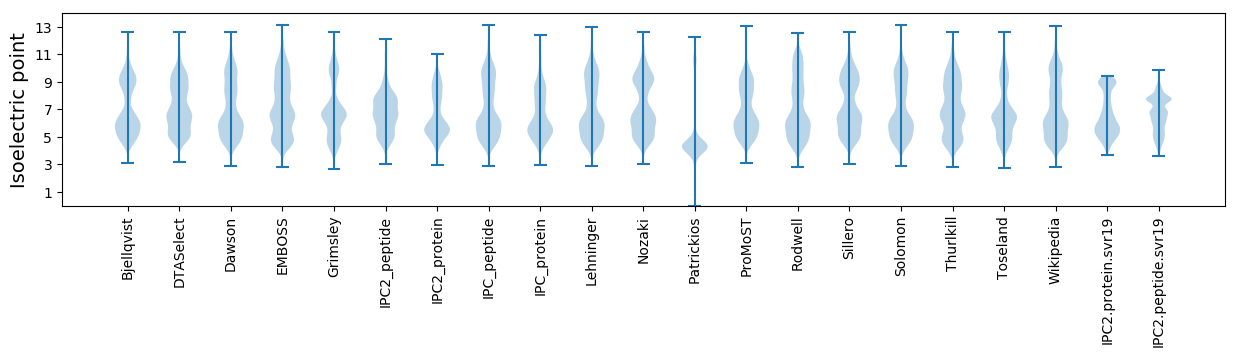
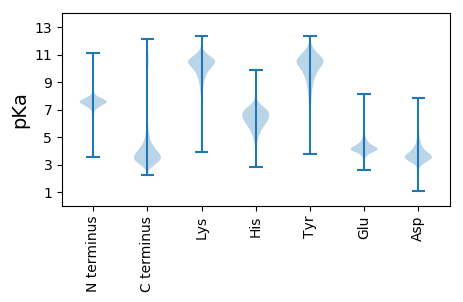
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1Y2DR73|A0A1Y2DR73_9PEZI Ketoacyl-synt_C domain-containing protein (Fragment) OS=Pseudomassariella vexata OX=1141098 GN=BCR38DRAFT_347936 PE=4 SV=1
MM1 pKa = 7.28PTLAPEE7 pKa = 4.29LTKK10 pKa = 10.7DD11 pKa = 3.86APDD14 pKa = 3.5LAKK17 pKa = 10.47RR18 pKa = 11.84AACTFSGADD27 pKa = 3.89GYY29 pKa = 11.93ASASKK34 pKa = 10.69SKK36 pKa = 10.64ADD38 pKa = 3.51CATIVLSALTVPSGVTLDD56 pKa = 4.05LTDD59 pKa = 4.84LSDD62 pKa = 3.6GTLVTFEE69 pKa = 5.81GEE71 pKa = 4.51TTWEE75 pKa = 4.09YY76 pKa = 11.34EE77 pKa = 3.98EE78 pKa = 4.54WEE80 pKa = 4.74GPLFAVSGTKK90 pKa = 8.58ITVDD94 pKa = 3.33GASGHH99 pKa = 5.65SLNGNGAKK107 pKa = 8.91WWDD110 pKa = 3.57GEE112 pKa = 4.23GDD114 pKa = 3.75SGVTKK119 pKa = 10.47PKK121 pKa = 9.94MFQAHH126 pKa = 6.69DD127 pKa = 5.16LIDD130 pKa = 3.66STIQNINIVNSPVQVFSINGCDD152 pKa = 3.27TLAVNSVTIDD162 pKa = 3.42NSDD165 pKa = 3.57GDD167 pKa = 4.53DD168 pKa = 4.38DD169 pKa = 5.19SLGHH173 pKa = 5.98NTDD176 pKa = 3.85CFDD179 pKa = 3.72IGSSTGVTITDD190 pKa = 3.72ATCYY194 pKa = 10.48NQDD197 pKa = 3.22DD198 pKa = 4.24CVAINSGTDD207 pKa = 2.85ITFTGGYY214 pKa = 9.4CSGGHH219 pKa = 5.94GLSIGSIGGRR229 pKa = 11.84DD230 pKa = 3.89DD231 pKa = 3.75NTVKK235 pKa = 10.08TVTFSDD241 pKa = 3.54SKK243 pKa = 10.9IINSQNGVRR252 pKa = 11.84IKK254 pKa = 9.89TKK256 pKa = 10.59SGDD259 pKa = 3.27TGTVEE264 pKa = 5.21DD265 pKa = 3.62ITYY268 pKa = 10.57RR269 pKa = 11.84DD270 pKa = 3.36ITLSGITDD278 pKa = 3.37YY279 pKa = 11.64GITVDD284 pKa = 4.03QAYY287 pKa = 10.59DD288 pKa = 3.54GDD290 pKa = 4.4DD291 pKa = 3.46ATDD294 pKa = 5.28GITISGFTLDD304 pKa = 4.56GVTGTTDD311 pKa = 2.62SDD313 pKa = 3.28ADD315 pKa = 3.49YY316 pKa = 10.93AIYY319 pKa = 10.41VNCGSDD325 pKa = 3.94SCTDD329 pKa = 3.4WTWTDD334 pKa = 3.4VSVTGADD341 pKa = 3.62NSCSNQPSGITCC353 pKa = 4.45
MM1 pKa = 7.28PTLAPEE7 pKa = 4.29LTKK10 pKa = 10.7DD11 pKa = 3.86APDD14 pKa = 3.5LAKK17 pKa = 10.47RR18 pKa = 11.84AACTFSGADD27 pKa = 3.89GYY29 pKa = 11.93ASASKK34 pKa = 10.69SKK36 pKa = 10.64ADD38 pKa = 3.51CATIVLSALTVPSGVTLDD56 pKa = 4.05LTDD59 pKa = 4.84LSDD62 pKa = 3.6GTLVTFEE69 pKa = 5.81GEE71 pKa = 4.51TTWEE75 pKa = 4.09YY76 pKa = 11.34EE77 pKa = 3.98EE78 pKa = 4.54WEE80 pKa = 4.74GPLFAVSGTKK90 pKa = 8.58ITVDD94 pKa = 3.33GASGHH99 pKa = 5.65SLNGNGAKK107 pKa = 8.91WWDD110 pKa = 3.57GEE112 pKa = 4.23GDD114 pKa = 3.75SGVTKK119 pKa = 10.47PKK121 pKa = 9.94MFQAHH126 pKa = 6.69DD127 pKa = 5.16LIDD130 pKa = 3.66STIQNINIVNSPVQVFSINGCDD152 pKa = 3.27TLAVNSVTIDD162 pKa = 3.42NSDD165 pKa = 3.57GDD167 pKa = 4.53DD168 pKa = 4.38DD169 pKa = 5.19SLGHH173 pKa = 5.98NTDD176 pKa = 3.85CFDD179 pKa = 3.72IGSSTGVTITDD190 pKa = 3.72ATCYY194 pKa = 10.48NQDD197 pKa = 3.22DD198 pKa = 4.24CVAINSGTDD207 pKa = 2.85ITFTGGYY214 pKa = 9.4CSGGHH219 pKa = 5.94GLSIGSIGGRR229 pKa = 11.84DD230 pKa = 3.89DD231 pKa = 3.75NTVKK235 pKa = 10.08TVTFSDD241 pKa = 3.54SKK243 pKa = 10.9IINSQNGVRR252 pKa = 11.84IKK254 pKa = 9.89TKK256 pKa = 10.59SGDD259 pKa = 3.27TGTVEE264 pKa = 5.21DD265 pKa = 3.62ITYY268 pKa = 10.57RR269 pKa = 11.84DD270 pKa = 3.36ITLSGITDD278 pKa = 3.37YY279 pKa = 11.64GITVDD284 pKa = 4.03QAYY287 pKa = 10.59DD288 pKa = 3.54GDD290 pKa = 4.4DD291 pKa = 3.46ATDD294 pKa = 5.28GITISGFTLDD304 pKa = 4.56GVTGTTDD311 pKa = 2.62SDD313 pKa = 3.28ADD315 pKa = 3.49YY316 pKa = 10.93AIYY319 pKa = 10.41VNCGSDD325 pKa = 3.94SCTDD329 pKa = 3.4WTWTDD334 pKa = 3.4VSVTGADD341 pKa = 3.62NSCSNQPSGITCC353 pKa = 4.45
Molecular weight: 36.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y2DFQ1|A0A1Y2DFQ1_9PEZI HET domain-containing protein OS=Pseudomassariella vexata OX=1141098 GN=BCR38DRAFT_448054 PE=4 SV=1
MM1 pKa = 7.32KK2 pKa = 9.55TRR4 pKa = 11.84KK5 pKa = 9.43KK6 pKa = 10.55SSPLPVSRR14 pKa = 11.84PRR16 pKa = 11.84QHH18 pKa = 6.69LAGPSRR24 pKa = 11.84SPITAPPTKK33 pKa = 9.38TRR35 pKa = 11.84KK36 pKa = 9.6KK37 pKa = 10.26SLPLPVSSPLATPNLASPKK56 pKa = 10.25
MM1 pKa = 7.32KK2 pKa = 9.55TRR4 pKa = 11.84KK5 pKa = 9.43KK6 pKa = 10.55SSPLPVSRR14 pKa = 11.84PRR16 pKa = 11.84QHH18 pKa = 6.69LAGPSRR24 pKa = 11.84SPITAPPTKK33 pKa = 9.38TRR35 pKa = 11.84KK36 pKa = 9.6KK37 pKa = 10.26SLPLPVSSPLATPNLASPKK56 pKa = 10.25
Molecular weight: 5.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5490982 |
49 |
4995 |
437.3 |
48.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.612 ± 0.02 | 1.312 ± 0.009 |
5.66 ± 0.016 | 5.968 ± 0.021 |
3.765 ± 0.013 | 7.075 ± 0.02 |
2.383 ± 0.009 | 4.93 ± 0.014 |
4.872 ± 0.02 | 8.804 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.007 | 3.772 ± 0.012 |
5.911 ± 0.024 | 3.927 ± 0.014 |
5.942 ± 0.019 | 8.099 ± 0.027 |
6.054 ± 0.016 | 6.262 ± 0.015 |
1.529 ± 0.009 | 2.805 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
